Crosstalk between mitochondrial stress signals regulates yeast chronological lifespan
- PMID: 24373996
- PMCID: PMC3943709
- DOI: 10.1016/j.mad.2013.12.002
Crosstalk between mitochondrial stress signals regulates yeast chronological lifespan
Abstract
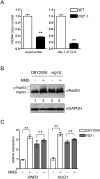
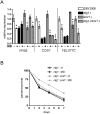
Mitochondrial DNA (mtDNA) exists in multiple copies per cell and is essential for oxidative phosphorylation. Depleted or mutated mtDNA promotes numerous human diseases and may contribute to aging. Reduced TORC1 signaling in the budding yeast, Saccharomyces cerevisiae, extends chronological lifespan (CLS) in part by generating a mitochondrial ROS (mtROS) signal that epigenetically alters nuclear gene expression. To address the potential requirement for mtDNA maintenance in this response, we analyzed strains lacking the mitochondrial base-excision repair enzyme Ntg1p. Extension of CLS by mtROS signaling and reduced TORC1 activity, but not caloric restriction, was abrogated in ntg1Δ strains that exhibited mtDNA depletion without defects in respiration. The DNA damage response (DDR) kinase Rad53p, which transduces pro-longevity mtROS signals, is also activated in ntg1Δ strains. Restoring mtDNA copy number alleviated Rad53p activation and re-established CLS extension following mtROS signaling, indicating that Rad53p senses mtDNA depletion directly. Finally, DDR kinases regulate nucleus-mitochondria localization dynamics of Ntg1p. From these results, we conclude that the DDR pathway senses and may regulate Ntg1p-dependent mtDNA stability. Furthermore, Rad53p senses multiple mitochondrial stresses in a hierarchical manner to elicit specific physiological outcomes, exemplified by mtDNA depletion overriding the ability of Rad53p to transduce an adaptive mtROS longevity signal.
Keywords: Chronological lifespan; DNA damage response; Rad53p; Reactive oxygen species; mtDNA.
Copyright © 2013 Elsevier Ireland Ltd. All rights reserved.
Figures



Similar articles
-
Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae.Mol Cell Biol. 2005 Jun;25(12):5196-204. doi: 10.1128/MCB.25.12.5196-5204.2005. Mol Cell Biol. 2005. PMID: 15923634 Free PMC article.
-
Epigenetic silencing mediates mitochondria stress-induced longevity.Cell Metab. 2013 Jun 4;17(6):954-964. doi: 10.1016/j.cmet.2013.04.003. Cell Metab. 2013. PMID: 23747251 Free PMC article.
-
Extension of chronological life span by reduced TOR signaling requires down-regulation of Sch9p and involves increased mitochondrial OXPHOS complex density.Aging (Albany NY). 2009 Jan 28;1(1):131-45. doi: 10.18632/aging.100016. Aging (Albany NY). 2009. PMID: 20157595 Free PMC article.
-
Regulation of longevity and oxidative stress by nutritional interventions: role of methionine restriction.Exp Gerontol. 2013 Oct;48(10):1030-42. doi: 10.1016/j.exger.2013.02.021. Epub 2013 Feb 27. Exp Gerontol. 2013. PMID: 23454735 Review.
-
Running on empty: does mitochondrial DNA mutation limit replicative lifespan in yeast?: Mutations that increase the division rate of cells lacking mitochondrial DNA also extend replicative lifespan in Saccharomyces cerevisiae.Bioessays. 2011 Oct;33(10):742-8. doi: 10.1002/bies.201100050. Epub 2011 Aug 9. Bioessays. 2011. PMID: 21826691 Review.
Cited by
-
FAM43A coordinates mtDNA replication and mitochondrial biogenesis in response to mtDNA depletion.J Cell Biol. 2025 Mar 3;224(3):e202311082. doi: 10.1083/jcb.202311082. Epub 2025 Jan 27. J Cell Biol. 2025. PMID: 39868925
-
Communications between Mitochondria, the Nucleus, Vacuoles, Peroxisomes, the Endoplasmic Reticulum, the Plasma Membrane, Lipid Droplets, and the Cytosol during Yeast Chronological Aging.Front Genet. 2016 Sep 27;7:177. doi: 10.3389/fgene.2016.00177. eCollection 2016. Front Genet. 2016. PMID: 27729926 Free PMC article. Review.
-
Mechanisms by which different functional states of mitochondria define yeast longevity.Int J Mol Sci. 2015 Mar 11;16(3):5528-54. doi: 10.3390/ijms16035528. Int J Mol Sci. 2015. PMID: 25768339 Free PMC article. Review.
-
Mitochondrial HMG-Box Containing Proteins: From Biochemical Properties to the Roles in Human Diseases.Biomolecules. 2020 Aug 16;10(8):1193. doi: 10.3390/biom10081193. Biomolecules. 2020. PMID: 32824374 Free PMC article. Review.
-
A Comprehensive Analysis of Replicative Lifespan in 4,698 Single-Gene Deletion Strains Uncovers Conserved Mechanisms of Aging.Cell Metab. 2015 Nov 3;22(5):895-906. doi: 10.1016/j.cmet.2015.09.008. Epub 2015 Oct 8. Cell Metab. 2015. PMID: 26456335 Free PMC article.
References
-
- Alseth I, Eide L, Pirovano M, Rognes T, Seeberg E, Bjoras M. The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Molecular and cellular biology. 1999;19:3779–3787. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

