Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets
- PMID: 24374217
- PMCID: PMC3932527
- DOI: 10.1016/j.cmet.2013.11.016
Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets
Abstract
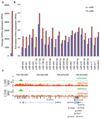
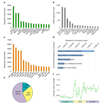
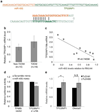
Type 2 diabetes mellitus (T2DM) is a complex disease characterized by the inability of the insulin-producing β cells in the endocrine pancreas to overcome insulin resistance in peripheral tissues. To determine if microRNAs are involved in the pathogenesis of human T2DM, we sequenced the small RNAs of human islets from diabetic and nondiabetic organ donors. We identified a cluster of microRNAs in an imprinted locus on human chromosome 14q32 that is highly and specifically expressed in human β cells and dramatically downregulated in islets from T2DM organ donors. The downregulation of this locus strongly correlates with hypermethylation of its promoter. Using HITS-CLIP for the essential RISC-component Argonaute, we identified disease-relevant targets of the chromosome 14q32 microRNAs, such as IAPP and TP53INP1, that cause increased β cell apoptosis upon overexpression in human islets. Our results support a role for microRNAs and their epigenetic control by DNA methylation in the pathogenesis of T2DM.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures



References
-
- Benetatos L, Hatzimichael E, Dasoula A, Dranitsaris G, Tsiara S, Syrrou M, Georgiou I, Bourantas KL. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leukemia research. 2010;34:148–153. - PubMed
-
- Bolmeson C, Esguerra JL, Salehi A, Speidel D, Eliasson L, Cilio CM. Differences in islet-enriched miRNAs in healthy and glucose intolerant human subjects. Biochemical and biophysical research communications. 2011;404:16–22. - PubMed
-
- Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52:102–110. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

