Identification of a major determinant for serine-threonine kinase phosphoacceptor specificity
- PMID: 24374310
- PMCID: PMC3898841
- DOI: 10.1016/j.molcel.2013.11.013
Identification of a major determinant for serine-threonine kinase phosphoacceptor specificity
Abstract
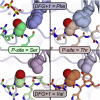
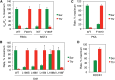
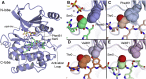
Eukaryotic protein kinases are generally classified as being either tyrosine or serine-threonine specific. Though not evident from inspection of their primary sequences, many serine-threonine kinases display a significant preference for serine or threonine as the phosphoacceptor residue. Here we show that a residue located in the kinase activation segment, which we term the "DFG+1" residue, acts as a major determinant for serine-threonine phosphorylation site specificity. Mutation of this residue was sufficient to switch the phosphorylation site preference for multiple kinases, including the serine-specific kinase PAK4 and the threonine-specific kinase MST4. Kinetic analysis of peptide substrate phosphorylation and crystal structures of PAK4-peptide complexes suggested that phosphoacceptor residue preference is not mediated by stronger binding of the favored substrate. Rather, favored kinase-phosphoacceptor combinations likely promote a conformation optimal for catalysis. Understanding the rules governing kinase phosphoacceptor preference allows kinases to be classified as serine or threonine specific based on their sequence.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Recognition of physiological phosphorylation sites by p21-activated kinase 4.J Struct Biol. 2020 Sep 1;211(3):107553. doi: 10.1016/j.jsb.2020.107553. Epub 2020 Jun 23. J Struct Biol. 2020. PMID: 32585314 Free PMC article.
-
Comprehensive profiling of the STE20 kinase family defines features essential for selective substrate targeting and signaling output.PLoS Biol. 2019 Mar 21;17(3):e2006540. doi: 10.1371/journal.pbio.2006540. eCollection 2019 Mar. PLoS Biol. 2019. PMID: 30897078 Free PMC article.
-
Acidophilic character of yeast PID261/BUD32, a putative ancestor of eukaryotic protein kinases.Biochem Biophys Res Commun. 2002 Sep 6;296(5):1366-71. doi: 10.1016/s0006-291x(02)02090-9. Biochem Biophys Res Commun. 2002. PMID: 12207926
-
Phosphorylation Sites in Protein Kinases and Phosphatases Regulated by Formyl Peptide Receptor 2 Signaling.Int J Mol Sci. 2020 May 27;21(11):3818. doi: 10.3390/ijms21113818. Int J Mol Sci. 2020. PMID: 32471307 Free PMC article. Review.
-
PDK2: the missing piece in the receptor tyrosine kinase signaling pathway puzzle.Am J Physiol Endocrinol Metab. 2005 Aug;289(2):E187-96. doi: 10.1152/ajpendo.00011.2005. Am J Physiol Endocrinol Metab. 2005. PMID: 16014356 Review.
Cited by
-
The C2 Domain and Altered ATP-Binding Loop Phosphorylation at Ser³⁵⁹ Mediate the Redox-Dependent Increase in Protein Kinase C-δ Activity.Mol Cell Biol. 2015 May;35(10):1727-40. doi: 10.1128/MCB.01436-14. Epub 2015 Mar 9. Mol Cell Biol. 2015. PMID: 25755284 Free PMC article.
-
Plasmodium falciparum Cyclic GMP-Dependent Protein Kinase Interacts with a Subunit of the Parasite Proteasome.Infect Immun. 2018 Dec 19;87(1):e00523-18. doi: 10.1128/IAI.00523-18. Print 2019 Jan. Infect Immun. 2018. PMID: 30323024 Free PMC article.
-
Dynamics of human protein kinase Aurora A linked to drug selectivity.Elife. 2018 Jun 14;7:e36656. doi: 10.7554/eLife.36656. Elife. 2018. PMID: 29901437 Free PMC article.
-
Identification and classification of small molecule kinases: insights into substrate recognition and specificity.BMC Evol Biol. 2016 Jan 6;16:7. doi: 10.1186/s12862-015-0576-x. BMC Evol Biol. 2016. PMID: 26738562 Free PMC article.
-
Self-assembly-based posttranslational protein oscillators.Sci Adv. 2020 Dec 16;6(51):eabc1939. doi: 10.1126/sciadv.abc1939. Print 2020 Dec. Sci Adv. 2020. PMID: 33328225 Free PMC article.
References
-
- Aimes R.T., Hemmer W., Taylor S.S. Serine-53 at the tip of the glycine-rich loop of cAMP-dependent protein kinase: role in catalysis, P-site specificity, and interaction with inhibitors. Biochemistry. 2000;39:8325–8332. - PubMed
-
- Brown N.R., Noble M.E., Endicott J.A., Johnson L.N. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1999;1:438–443. - PubMed
-
- Bullock A.N., Debreczeni J., Amos A.L., Knapp S., Turk B.E. Structure and substrate specificity of the Pim-1 kinase. J. Biol. Chem. 2005;280:41675–41682. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

