The versatility of the αβ T-cell antigen receptor
- PMID: 24375592
- PMCID: PMC3945834
- DOI: 10.1002/pro.2412
The versatility of the αβ T-cell antigen receptor
Abstract
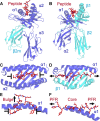
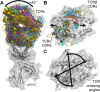
The T-cell antigen receptor is a heterodimeric αβ protein (TCR) expressed on the surface of T-lymphocytes, with each chain of the TCR comprising three complementarity-determining regions (CDRs) that collectively form the antigen-binding site. Unlike antibodies, which are closely related proteins that recognize intact protein antigens, TCRs classically bind, via their CDR loops, to peptides (p) that are presented by molecules of the major histocompatibility complex (MHC). This TCR-pMHC interaction is crucially important in cell-mediated immunity, with the specificity in the cellular immune response being attributable to MHC polymorphism, an extensive TCR repertoire and a variable peptide cargo. The ensuing structural and biophysical studies within the TCR-pMHC axis have been highly informative in understanding the fundamental events that underpin protective immunity and dysfunctional T-cell responses that occur during autoimmunity. In addition, TCRs can recognize the CD1 family, a family of MHC-related molecules that instead of presenting peptides are ideally suited to bind lipid-based antigens. Structural studies within the CD1-lipid antigen system are beginning to inform us how lipid antigens are specifically presented by CD1, and how such CD1-lipid antigen complexes are recognized by the TCR. Moreover, it has recently been shown that certain TCRs can bind to vitamin B based metabolites that are bound to an MHC-like molecule termed MR1. Thus, TCRs can recognize peptides, lipids, and small molecule metabolites, and here we review the basic principles underpinning this versatile and fascinating receptor recognition system that is vital to a host's survival.
Keywords: T-cell receptor; antigen presenting molecules; immune system; structures.
© 2013 The Protein Society.
Figures

References
-
- Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of T-cell receptor bias in immunity. Nat Rev Immunol. 2006;6:883–894. - PubMed
-
- Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. - PubMed
-
- Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA. An αβ T-cell receptor structure at 2.5 Å and its orientation in the TCR-MHC complex. Science. 1996;274:209–219. - PubMed
-
- Berry R, Chen Z, McCluskey J, Rossjohn J. Insight into the basis of autonomous immunoreceptor activation. Trends Immunol. 2011;32:165–170. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

