Ubiquitin in the immune system
- PMID: 24375678
- PMCID: PMC4303447
- DOI: 10.1002/embr.201338025
Ubiquitin in the immune system
Erratum in
- EMBO Rep. 2014 Mar;15(3):322
Abstract
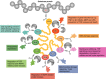
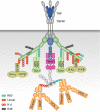
Ubiquitination is a post-translational modification process that has been implicated in the regulation of innate and adaptive immune responses. There is increasing evidence that both ubiquitination and its reversal, deubiquitination, play crucial roles not only during the development of the immune system but also in the orchestration of an immune response by ensuring the proper functioning of the different cell types that constitute the immune system. Here, we provide an overview of the latest discoveries in this field and discuss how they impact our understanding of the ubiquitin system in host defence mechanisms as well as self-tolerance.
Figures


References
-
- Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. - PubMed
-
- Ciechanover A, Elias S, Heller H, Hershko A. ‘Covalent affinity’ purification of ubiquitin-activating enzyme. J Biol Chem. 1982;257:2537–2542. - PubMed
-
- Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–8214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

