Use of a phosphonate methyltransferase in the identification of the fosfazinomycin biosynthetic gene cluster
- PMID: 24376039
- PMCID: PMC3927463
- DOI: 10.1002/anie.201308363
Use of a phosphonate methyltransferase in the identification of the fosfazinomycin biosynthetic gene cluster
Abstract
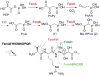
Natural product discovery has been boosted by genome mining approaches, but compound purification is often still challenging. We report an enzymatic strategy for "stable isotope labeling of phosphonates in extract" (SILPE) that facilitates their purification. We used the phosphonate methyltransferase DhpI involved in dehydrophos biosynthesis to methylate a variety of phosphonate natural products in crude spent medium with a mixture of labeled and unlabeled S-adenosyl methionine. Mass-guided fractionation then allowed straightforward purification. We illustrate its utility by purifying a phosphonate that led to the identification of the fosfazinomycin biosynthetic gene cluster. This unusual natural product contains a hydrazide linker between a carboxylic acid and a phosphonic acid. Bioinformatic analysis of the gene cluster provides insights into how such a structure might be assembled.
Keywords: antibiotics; biosynthesis; hydrazine; natural products.
Copyright © 2014 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures



References
-
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Mol Cell Proteom. 2002;1:376–386. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

