IRES-mediated translation of membrane proteins and glycoproteins in eukaryotic cell-free systems
- PMID: 24376523
- PMCID: PMC3869664
- DOI: 10.1371/journal.pone.0082234
IRES-mediated translation of membrane proteins and glycoproteins in eukaryotic cell-free systems
Abstract
Internal ribosome entry site (IRES) elements found in the 5' untranslated region of mRNAs enable translation initiation in a cap-independent manner, thereby representing an alternative to cap-dependent translation in cell-free protein expression systems. However, IRES function is largely species-dependent so their utility in cell-free systems from different species is rather limited. A promising approach to overcome these limitations would be the use of IRESs that are able to recruit components of the translation initiation apparatus from diverse origins. Here, we present a solution to this technical problem and describe the ability of a number of viral IRESs to direct efficient protein expression in different eukaryotic cell-free expression systems. The IRES from the intergenic region (IGR) of the Cricket paralysis virus (CrPV) genome was shown to function efficiently in four different cell-free systems based on lysates derived from cultured Sf21, CHO and K562 cells as well as wheat germ. Our results suggest that the CrPV IGR IRES-based expression vector is universally applicable for a broad range of eukaryotic cell lysates. Sf21, CHO and K562 cell-free expression systems are particularly promising platforms for the production of glycoproteins and membrane proteins since they contain endogenous microsomes that facilitate the incorporation of membrane-spanning proteins and the formation of post-translational modifications. We demonstrate the use of the CrPV IGR IRES-based expression vector for the enhanced synthesis of various target proteins including the glycoprotein erythropoietin and the membrane proteins heparin-binding EGF-like growth factor receptor as well as epidermal growth factor receptor in the above mentioned eukaryotic cell-free systems. CrPV IGR IRES-mediated translation will facilitate the development of novel eukaryotic cell-free expression platforms as well as the high-yield synthesis of desired proteins in already established systems.
Conflict of interest statement
Figures
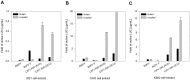
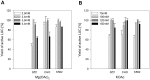




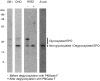
References
-
- Marintchev A, Wagner G (2004) Translation initiation: Structures, mechanisms and evolution. Quarterly Reviews of Biophysics 37: 197–284. - PubMed
-
- Swartz JR (2009) Universal cell-free protein synthesis. Nat Biotech 27: 731–732. - PubMed
-
- Lodish H, Berk A, Zipursky S (2000) Processing of eukaryotic mRNA. Molecular Cell Biology 4th edition New York: W H Freeman; 2000 Section 112.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

