The PCNA interaction protein box sequence in Rad54 is an integral part of its ATPase domain and is required for efficient DNA repair and recombination
- PMID: 24376557
- PMCID: PMC3869717
- DOI: 10.1371/journal.pone.0082630
The PCNA interaction protein box sequence in Rad54 is an integral part of its ATPase domain and is required for efficient DNA repair and recombination
Erratum in
- PLoS One. 2014;9(6):e101095
Abstract
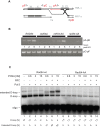
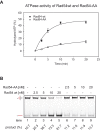
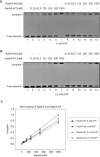
Rad54 is an ATP-driven translocase involved in the genome maintenance pathway of homologous recombination (HR). Although its activity has been implicated in several steps of HR, its exact role(s) at each step are still not fully understood. We have identified a new interaction between Rad54 and the replicative DNA clamp, proliferating cell nuclear antigen (PCNA). This interaction was only mildly weakened by the mutation of two key hydrophobic residues in the highly-conserved PCNA interaction motif (PIP-box) of Rad54 (Rad54-AA). Intriguingly, the rad54-AA mutant cells displayed sensitivity to DNA damage and showed HR defects similar to the null mutant, despite retaining its ability to interact with HR proteins and to be recruited to HR foci in vivo. We therefore surmised that the PCNA interaction might be impaired in vivo and was unable to promote repair synthesis during HR. Indeed, the Rad54-AA mutant was defective in primer extension at the MAT locus as well as in vitro, but additional biochemical analysis revealed that this mutant also had diminished ATPase activity and an inability to promote D-loop formation. Further mutational analysis of the putative PIP-box uncovered that other phenotypically relevant mutants in this domain also resulted in a loss of ATPase activity. Therefore, we have found that although Rad54 interacts with PCNA, the PIP-box motif likely plays only a minor role in stabilizing the PCNA interaction, and rather, this conserved domain is probably an extension of the ATPase domain III.
Conflict of interest statement
Figures


Similar articles
-
A conserved sequence extending motif III of the motor domain in the Snf2-family DNA translocase Rad54 is critical for ATPase activity.PLoS One. 2013 Dec 16;8(12):e82184. doi: 10.1371/journal.pone.0082184. eCollection 2013. PLoS One. 2013. PMID: 24358152 Free PMC article.
-
Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination.Biochim Biophys Acta. 2011 Sep;1809(9):509-23. doi: 10.1016/j.bbagrm.2011.06.006. Epub 2011 Jun 16. Biochim Biophys Acta. 2011. PMID: 21704205 Free PMC article. Review.
-
Effects of tumor-associated mutations on Rad54 functions.J Biol Chem. 2004 Jun 4;279(23):24081-8. doi: 10.1074/jbc.M402719200. Epub 2004 Mar 31. J Biol Chem. 2004. PMID: 15056673
-
In vitro role of Rad54 in Rad51-ssDNA filament-dependent homology search and synaptic complexes formation.Nat Commun. 2019 Sep 6;10(1):4058. doi: 10.1038/s41467-019-12082-z. Nat Commun. 2019. PMID: 31492866 Free PMC article.
-
Rad54: the Swiss Army knife of homologous recombination?Nucleic Acids Res. 2006;34(15):4115-25. doi: 10.1093/nar/gkl481. Epub 2006 Aug 25. Nucleic Acids Res. 2006. PMID: 16935872 Free PMC article. Review.
Cited by
-
Geminivirus Replication Protein Impairs SUMO Conjugation of Proliferating Cellular Nuclear Antigen at Two Acceptor Sites.J Virol. 2018 Aug 29;92(18):e00611-18. doi: 10.1128/JVI.00611-18. Print 2018 Sep 15. J Virol. 2018. PMID: 29950424 Free PMC article.
-
A residue of motif III positions the helicase domains of motor subunit HsdR in restriction-modification enzyme EcoR124I.J Mol Model. 2018 Jun 26;24(7):176. doi: 10.1007/s00894-018-3722-8. J Mol Model. 2018. PMID: 29943199
-
Mutations at the Subunit Interface of Yeast Proliferating Cell Nuclear Antigen Reveal a Versatile Regulatory Domain.PLoS One. 2016 Aug 18;11(8):e0161307. doi: 10.1371/journal.pone.0161307. eCollection 2016. PLoS One. 2016. PMID: 27537501 Free PMC article.
-
Physiology of the read-write genome.J Physiol. 2014 Jun 1;592(11):2319-41. doi: 10.1113/jphysiol.2014.271130. J Physiol. 2014. PMID: 24882816 Free PMC article. Review.
-
Characterization of proliferating cell nuclear antigen (PCNA) from pathogenic yeast Candida albicans and its functional analyses in S. cerevisiae.BMC Microbiol. 2015 Nov 4;15:257. doi: 10.1186/s12866-015-0582-6. BMC Microbiol. 2015. PMID: 26537947 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

