Next-generation sequencing of lung cancer EGFR exons 18-21 allows effective molecular diagnosis of small routine samples (cytology and biopsy)
- PMID: 24376723
- PMCID: PMC3871524
- DOI: 10.1371/journal.pone.0083607
Next-generation sequencing of lung cancer EGFR exons 18-21 allows effective molecular diagnosis of small routine samples (cytology and biopsy)
Abstract
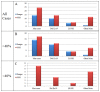
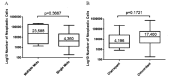
Selection of lung cancer patients for therapy with tyrosine kinase inhibitors directed at EGFR requires the identification of specific EGFR mutations. In most patients with advanced, inoperable lung carcinoma limited tumor samples often represent the only material available for both histologic typing and molecular analysis. We defined a next generation sequencing protocol targeted to EGFR exons 18-21 suitable for the routine diagnosis of such clinical samples. The protocol was validated in an unselected series of 80 small biopsies (n=14) and cytology (n=66) specimens representative of the material ordinarily submitted for diagnostic evaluation to three referral medical centers in Italy. Specimens were systematically evaluated for tumor cell number and proportion relative to non-neoplastic cells. They were analyzed in batches of 100-150 amplicons per run, reaching an analytical sensitivity of 1% and obtaining an adequate number of reads, to cover all exons on all samples analyzed. Next generation sequencing was compared with Sanger sequencing. The latter identified 15 EGFR mutations in 14/80 cases (17.5%) but did not detected mutations when the proportion of neoplastic cells was below 40%. Next generation sequencing identified 31 EGFR mutations in 24/80 cases (30.0%). Mutations were detected with a proportion of neoplastic cells as low as 5%. All mutations identified by the Sanger method were confirmed. In 6 cases next generation sequencing identified exon 19 deletions or the L858R mutation not seen after Sanger sequencing, allowing the patient to be treated with tyrosine kinase inhibitors. In one additional case the R831H mutation associated with treatment resistance was identified in an EGFR wild type tumor after Sanger sequencing. Next generation sequencing is robust, cost-effective and greatly improves the detection of EGFR mutations. Its use should be promoted for the clinical diagnosis of mutations in specimens with unfavorable tumor cell content.
Conflict of interest statement
Figures




References
-
- Fukuoka M, Wu YL, Thongprasert S, Sunpaweravong P, Leong SS et al. (2011) Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol 29: 2866-2874. doi: 10.1200/JCO.2010.33.4235. PubMed: 21670455. - DOI - PubMed
-
- Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I et al. (2010) Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 11: 121-128. doi: 10.1016/S1470-2045(09)70364-X. PubMed: 20022809. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

