Differentially expressed wound healing-related microRNAs in the human diabetic cornea
- PMID: 24376808
- PMCID: PMC3869828
- DOI: 10.1371/journal.pone.0084425
Differentially expressed wound healing-related microRNAs in the human diabetic cornea
Abstract
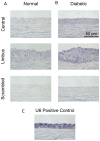
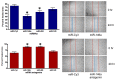
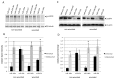
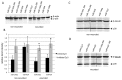
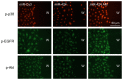
MicroRNAs are powerful gene expression regulators, but their corneal repertoire and potential changes in corneal diseases remain unknown. Our purpose was to identify miRNAs altered in the human diabetic cornea by microarray analysis, and to examine their effects on wound healing in cultured telomerase-immortalized human corneal epithelial cells (HCEC) in vitro. Total RNA was extracted from age-matched human autopsy normal (n=6) and diabetic (n=6) central corneas, Flash Tag end-labeled, and hybridized to Affymetrix® GeneChip® miRNA Arrays. Select miRNAs associated with diabetic cornea were validated by quantitative RT-PCR (Q-PCR) and by in situ hybridization (ISH) in independent samples. HCEC were transfected with human pre-miR™miRNA precursors (h-miR) or their inhibitors (antagomirs) using Lipofectamine 2000. Confluent transfected cultures were scratch-wounded with P200 pipette tip. Wound closure was monitored by digital photography. Expression of signaling proteins was detected by immunostaining and Western blot. Using microarrays, 29 miRNAs were identified as differentially expressed in diabetic samples. Two miRNA candidates showing the highest fold increased in expression in the diabetic cornea were confirmed by Q-PCR and further characterized. HCEC transfection with h-miR-146a or h-miR-424 significantly retarded wound closure, but their respective antagomirs significantly enhanced wound healing vs. controls. Cells treated with h-miR-146a or h-miR-424 had decreased p-p38 and p-EGFR staining, but these increased over control levels close to the wound edge upon antagomir treatment. In conclusion, several miRNAs with increased expression in human diabetic central corneas were found. Two such miRNAs inhibited cultured corneal epithelial cell wound healing. Dysregulation of miRNA expression in human diabetic cornea may be an important mediator of abnormal wound healing.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

