Gene expression analysis indicates CB1 receptor upregulation in the hippocampus and neurotoxic effects in the frontal cortex 3 weeks after single-dose MDMA administration in Dark Agouti rats
- PMID: 24378229
- PMCID: PMC3902429
- DOI: 10.1186/1471-2164-14-930
Gene expression analysis indicates CB1 receptor upregulation in the hippocampus and neurotoxic effects in the frontal cortex 3 weeks after single-dose MDMA administration in Dark Agouti rats
Retraction in
-
Retraction Note to: Gene expression analysis indicates CB1 receptor upregulation in the hippocampus and neurotoxic effects in the frontal cortex 3 weeks after single-dose MDMA administration in Dark Agouti rats.BMC Genomics. 2016 Sep 8;17(1):721. doi: 10.1186/s12864-016-3072-9. BMC Genomics. 2016. PMID: 27608804 Free PMC article. No abstract available.
Abstract
Background: 3,4-methylenedioxymethamphetamine (MDMA, "ecstasy") is a widely used recreational drug known to impair cognitive functions on the long-run. Both hippocampal and frontal cortical regions have well established roles in behavior, memory formation and other cognitive tasks and damage of these regions is associated with altered behavior and cognitive functions, impairments frequently described in heavy MDMA users. The aim of this study was to examine the hippocampus, frontal cortex and dorsal raphe of Dark Agouti rats with gene expression arrays (Illumina RatRef bead arrays) looking for possible mechanisms and new candidates contributing to the effects of a single dose of MDMA (15 mg/kg) 3 weeks earlier.
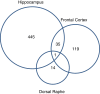
Results: The number of differentially expressed genes in the hippocampus, frontal cortex and the dorsal raphe were 481, 155, and 15, respectively. Gene set enrichment analysis of the microarray data revealed reduced expression of 'memory' and 'cognition', 'dendrite development' and 'regulation of synaptic plasticity' gene sets in the hippocampus, parallel to the upregulation of the CB1 cannabinoid- and Epha4, Epha5, Epha6 ephrin receptors. Downregulated gene sets in the frontal cortex were related to protein synthesis, chromatin organization, transmembrane transport processes, while 'dendrite development', 'regulation of synaptic plasticity' and 'positive regulation of synapse assembly' gene sets were upregulated. Changes in the dorsal raphe region were mild and in most cases not significant.
Conclusion: The present data raise the possibility of new synapse formation/synaptic reorganization in the frontal cortex three weeks after a single neurotoxic dose of MDMA. In contrast, a prolonged depression of new neurite formation in the hippocampus is suggested by the data, which underlines the particular vulnerability of this brain region after the drug treatment. Finally, our results also suggest the substantial contribution of CB1 receptor and endocannabinoid mediated pathways in the hippocampal impairments. Taken together the present study provides evidence for the participation of new molecular candidates in the long-term effects of MDMA.
Figures





Similar articles
-
Gene expression analysis indicates reduced memory and cognitive functions in the hippocampus and increase in synaptic reorganization in the frontal cortex 3 weeks after MDMA administration in Dark Agouti rats.BMC Genomics. 2018 Aug 2;19(1):580. doi: 10.1186/s12864-018-4929-x. BMC Genomics. 2018. PMID: 30071829 Free PMC article.
-
Active and passive MDMA ('ecstasy') intake induces differential transcriptional changes in the mouse brain.Genes Brain Behav. 2012 Feb;11(1):38-51. doi: 10.1111/j.1601-183X.2011.00735.x. Epub 2011 Nov 16. Genes Brain Behav. 2012. PMID: 21951708
-
Increased CRE-binding activity and tryptophan hydroxylase mRNA expression induced by 3,4-methylenedioxymethamphetamine (MDMA, "ecstasy") in the rat frontal cortex but not in the hippocampus.Brain Res Mol Brain Res. 2004 Jul 26;126(2):181-7. doi: 10.1016/j.molbrainres.2004.04.006. Brain Res Mol Brain Res. 2004. PMID: 15249142
-
Elevated BDNF protein level in cortex but not in hippocampus of MDMA-treated Dark Agouti rats: a potential link to the long-term recovery of serotonergic axons.Neurosci Lett. 2010 Jul 5;478(2):56-60. doi: 10.1016/j.neulet.2010.04.061. Epub 2010 May 8. Neurosci Lett. 2010. PMID: 20435092
-
The effects of methylenedioxymethamphetamine (MDMA, "Ecstasy") on monoaminergic neurotransmission in the central nervous system.Prog Neurobiol. 1996 Aug;49(5):455-79. doi: 10.1016/0301-0082(96)00027-5. Prog Neurobiol. 1996. PMID: 8895996 Review.
Cited by
-
Retraction Note to: Gene expression analysis indicates CB1 receptor upregulation in the hippocampus and neurotoxic effects in the frontal cortex 3 weeks after single-dose MDMA administration in Dark Agouti rats.BMC Genomics. 2016 Sep 8;17(1):721. doi: 10.1186/s12864-016-3072-9. BMC Genomics. 2016. PMID: 27608804 Free PMC article. No abstract available.
-
Time-course adaptive changes in hippocampal transcriptome and synaptic function induced by simulated microgravity associated with cognition.Front Cell Neurosci. 2023 Oct 5;17:1275771. doi: 10.3389/fncel.2023.1275771. eCollection 2023. Front Cell Neurosci. 2023. PMID: 37868195 Free PMC article.
-
Transcriptional evidence for the role of chronic venlafaxine treatment in neurotrophic signaling and neuroplasticity including also Glutamatergic [corrected] - and insulin-mediated neuronal processes.PLoS One. 2014 Nov 25;9(11):e113662. doi: 10.1371/journal.pone.0113662. eCollection 2014. PLoS One. 2014. PMID: 25423262 Free PMC article.
References
-
- Colado MI, O’Shea E, Granados R, Esteban B, Martin AB, Green AR. Studies on the role of dopamine in the degeneration of 5-HT nerve endings in the brain of Dark Agouti rats following 3,4-methylenedioxymethamphetamine (MDMA or 'ecstasy’) administration. Br J Pharmacol. 1999;14(4):911–924. doi: 10.1038/sj.bjp.0702373. - DOI - PMC - PubMed
-
- Adori C, Ando RD, Kovacs GG, Bagdy G. Damage of serotonergic axons and immunolocalization of Hsp27, Hsp72, and Hsp90 molecular chaperones after a single dose of MDMA administration in Dark Agouti rat: temporal, spatial, and cellular patterns. J Comp Neurol. 2006;14(2):251–269. doi: 10.1002/cne.20994. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

