Replication cycle and molecular biology of the West Nile virus
- PMID: 24378320
- PMCID: PMC3917430
- DOI: 10.3390/v6010013
Replication cycle and molecular biology of the West Nile virus
Abstract
West Nile virus (WNV) is a member of the genus Flavivirus in the family Flaviviridae. Flaviviruses replicate in the cytoplasm of infected cells and modify the host cell environment. Although much has been learned about virion structure and virion-endosomal membrane fusion, the cell receptor(s) used have not been definitively identified and little is known about the early stages of the virus replication cycle. Members of the genus Flavivirus differ from members of the two other genera of the family by the lack of a genomic internal ribosomal entry sequence and the creation of invaginations in the ER membrane rather than double-membrane vesicles that are used as the sites of exponential genome synthesis. The WNV genome 3' and 5' sequences that form the long distance RNA-RNA interaction required for minus strand initiation have been identified and contact sites on the 5' RNA stem loop for NS5 have been mapped. Structures obtained for many of the viral proteins have provided information relevant to their functions. Viral nonstructural protein interactions are complex and some may occur only in infected cells. Although interactions between many cellular proteins and virus components have been identified, the functions of most of these interactions have not been delineated.
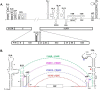
Figures

References
-
- Berthet F.X., Zeller H.G., Drouet M.T., Rauzier J., Digoutte J.P., Deubel V. Extensive nucleotide changes and deletions within the envelope glycoprotein gene of Euro-African West Nile viruses. J. Gen. Virol. 1997;78:2293–2297. - PubMed
-
- Lanciotti R.S., Roehrig J.T., Deubel V., Smith J., Parker M., Steele K., Crise B., Volpe K.E., Crabtree M.B., Scherret J.H., et al. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science. 1999;286:2333–2337. doi: 10.1126/science.286.5448.2333. - DOI - PubMed
-
- Heinz F.X., Purcell M.S., Gould E.A., Howard C.R., Houghton M., Moormann R.J.M., Rice C.M., Thiel H.-J. Family Flaviviridae. In: Regenmortel C.F., Bishop D.H.L., Carstens E.B., Estes M.K., editors. Virus Taxonomy. Academic Press; San Diego, CA, USA: 2000. pp. 860–878.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

