The Role of the 3' Untranslated Region in the Post-Transcriptional Regulation of KLF6 Gene Expression in Hepatocellular Carcinoma
- PMID: 24378751
- PMCID: PMC3980593
- DOI: 10.3390/cancers6010028
The Role of the 3' Untranslated Region in the Post-Transcriptional Regulation of KLF6 Gene Expression in Hepatocellular Carcinoma
Abstract
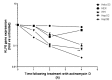
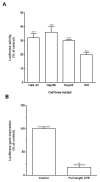
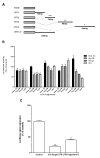
KLF6 is ubiquitously expressed in human tissues and regulates many pathways such as differentiation, development, cellular proliferation, growth-related signal transduction, and apoptosis. We previously demonstrated that KLF6 expression is altered during liver carcinogenesis. More importantly, KLF6 invalidation results in cell cycle progression inhibition and apoptosis of liver cancer cells. On the other hand, enforced expression of KLF6 variant 2 (SV2) induces cancer cell death by apoptosis. Thus, we and others demonstrated that KLF6 and its splicing variants play a critical role in liver cancer. However, little is known on the mechanisms governing KLF6 expression in HCC. In the present work, we asked whether the 3' untranslated region (3'UTR) of the KLF6 mRNA may be responsible for regulation of KLF6 expression in HCC. We found that KLF6 mRNA stability was altered in liver-derived cell lines as compared to cervical cancer-derived cell lines and human embryonic fibroblasts. Interestingly, KLF6 mRNA was highly unstable in liver cancer-derived cell lines as compared to normal hepatocytes. We next cloned the KLF6 mRNA 3'UTR into luciferase-expressing vectors and found that gene expression and activity were strongly impaired in all liver-derived cell lines tested. In addition, we found that most the KLF6 3'UTR destabilisation activity resides between nt 1,835 and nt 2,615 of the KLF6 gene. Taken together, we provide the first steps towards better understanding of the regulation of KLF6 expression in HCC. Further work is needed to identify the factors that bind to KLF6 3'UTR to regulate its expression in liver cancer-derived cell lines.
Figures

References
-
- Benzeno S., Narla G., Allina J., Cheng G.Z., Reeves H.L., Banck M.S., Odin J.A., Diehl J.A., Germain D., Friedman S.L. Cyclin-dependent kinase inhibition by the KLF6 tumor suppressor protein through interaction with cyclin D1. Cancer Res. 2004;64:3885–3891. doi: 10.1158/0008-5472.CAN-03-2818. - DOI - PubMed
-
- Kremer-Tal S., Reeves H.L., Narla G., Thung S.N., Schwartz M., Difeo A., Katz A., Bruix J., Bioulac-Sage P., Martignetti J.A., et al. Frequent inactivation of the tumor suppressor Kruppel-like factor 6 (KLF6) in hepatocellular carcinoma. Hepatology. 2004;40:1047–1052. doi: 10.1002/hep.20460. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

