Genetic variation in the green anole lizard (Anolis carolinensis) reveals island refugia and a fragmented Florida during the quaternary
- PMID: 24379168
- PMCID: PMC4778398
- DOI: 10.1007/s10709-013-9754-1
Genetic variation in the green anole lizard (Anolis carolinensis) reveals island refugia and a fragmented Florida during the quaternary
Abstract
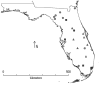
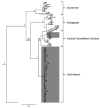
The green anole lizard (Anolis carolinensis) is a model organism for behavior and genomics that is native to the southeastern United States. It is currently thought that the ancestors of modern green anoles dispersed to peninsular Florida from Cuba. However, the climatic changes and geological features responsible for the early diversification of A. carolinensis in North America have remained largely unexplored. This is because previous studies (1) differ in their estimates of the divergence times of populations, (2) are based on a single genetic locus or (3) did not test specific hypotheses regarding the geologic and topographic history of Florida. Here we provide a multi-locus study of green anole genetic diversity and find that the Florida peninsula contains a larger number of genetically distinct populations that are more diverse than those on the continental mainland. As a test of the island refugia hypothesis in Pleistocene Florida, we use a coalescent approach to estimate the divergence times of modern green anole lineages. We find that all demographic events occurred during or after the Upper Pliocene and suggest that green anole diversification was driven by population divergence on interglacial island refugia in Florida during the Lower Pleistocene, while the region was often separated from continental North America. When Florida reconnected to the mainland, two separate dispersal events led to the expansion of green anole populations across the Atlantic Seaboard and Gulf Coastal Plain.
Figures




Similar articles
-
Multi-locus phylogeographic and population genetic analysis of Anolis carolinensis: historical demography of a genomic model species.PLoS One. 2012;7(6):e38474. doi: 10.1371/journal.pone.0038474. Epub 2012 Jun 7. PLoS One. 2012. PMID: 22685573 Free PMC article.
-
Out of Florida: mtDNA reveals patterns of migration and Pleistocene range expansion of the Green Anole lizard (Anolis carolinensis).Ecol Evol. 2012 Sep;2(9):2274-84. doi: 10.1002/ece3.324. Epub 2012 Aug 8. Ecol Evol. 2012. PMID: 23139885 Free PMC article.
-
Hybridization and rapid differentiation after secondary contact between the native green anole (Anolis carolinensis) and the introduced green anole (Anolis porcatus).Ecol Evol. 2019 Mar 26;9(7):4138-4148. doi: 10.1002/ece3.5042. eCollection 2019 Apr. Ecol Evol. 2019. PMID: 31015994 Free PMC article.
-
Out of Cuba: overwater dispersal and speciation among lizards in the Anolis carolinensis subgroup.Mol Ecol. 2005 Jul;14(8):2419-32. doi: 10.1111/j.1365-294X.2005.02550.x. Mol Ecol. 2005. PMID: 15969724
-
Recent Secondary Contacts, Linked Selection, and Variable Recombination Rates Shape Genomic Diversity in the Model Species Anolis carolinensis.Genome Biol Evol. 2019 Jul 1;11(7):2009-2022. doi: 10.1093/gbe/evz110. Genome Biol Evol. 2019. PMID: 31134281 Free PMC article.
Cited by
-
Diversification in wild populations of the model organism Anolis carolinensis: A genome-wide phylogeographic investigation.Ecol Evol. 2016 Oct 17;6(22):8115-8125. doi: 10.1002/ece3.2547. eCollection 2016 Nov. Ecol Evol. 2016. PMID: 27891220 Free PMC article.
-
Barcoding blood meals: New vertebrate-specific primer sets for assigning taxonomic identities to host DNA from mosquito blood meals.PLoS Negl Trop Dis. 2018 Aug 30;12(8):e0006767. doi: 10.1371/journal.pntd.0006767. eCollection 2018 Aug. PLoS Negl Trop Dis. 2018. PMID: 30161128 Free PMC article.
-
Chromosome-scale genome assembly of the brown anole (Anolis sagrei), an emerging model species.Commun Biol. 2022 Oct 25;5(1):1126. doi: 10.1038/s42003-022-04074-5. Commun Biol. 2022. PMID: 36284162 Free PMC article.
-
Disentangling the determinants of transposable elements dynamics in vertebrate genomes using empirical evidences and simulations.PLoS Genet. 2020 Oct 5;16(10):e1009082. doi: 10.1371/journal.pgen.1009082. eCollection 2020 Oct. PLoS Genet. 2020. PMID: 33017388 Free PMC article.
-
Comparative Genomics Reveals Accelerated Evolution in Conserved Pathways during the Diversification of Anole Lizards.Genome Biol Evol. 2018 Feb 1;10(2):489-506. doi: 10.1093/gbe/evy013. Genome Biol Evol. 2018. PMID: 29360978 Free PMC article.
References
-
- Bishop DC, Echternacht AC. Emergence behavior and movements of winter-aggregated green anoles (Anolis carolinensis) and the thermal characteristics of their crevices in Tennessee. Herpetologica. 2004;60 (2):168–177. doi: 10.1655/02-34. - DOI
-
- Buth DG, Gorman GC, Lieb CS. Genetic divergence between Anolis carolinensis and its Cuban progenitor, Anolis porcatus. Journal of Herpetology. 1980;14(3):279–284.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

