Eosinophil deficiency compromises lung defense against Aspergillus fumigatus
- PMID: 24379296
- PMCID: PMC3958002
- DOI: 10.1128/IAI.01172-13
Eosinophil deficiency compromises lung defense against Aspergillus fumigatus
Abstract
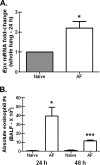
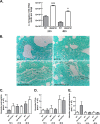
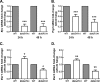
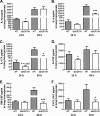
Exposure to the mold Aspergillus fumigatus may result in allergic bronchopulmonary aspergillosis, chronic necrotizing pulmonary aspergillosis, or invasive aspergillosis (IA), depending on the host's immune status. Neutrophil deficiency is the predominant risk factor for the development of IA, the most life-threatening condition associated with A. fumigatus exposure. Here we demonstrate that in addition to neutrophils, eosinophils are an important contributor to the clearance of A. fumigatus from the lung. Acute A. fumigatus challenge in normal mice induced the recruitment of CD11b+ Siglec F+ Ly-6G(lo) Ly-6C(neg) CCR3+ eosinophils to the lungs, which was accompanied by an increase in lung Epx (eosinophil peroxidase) mRNA levels. Mice deficient in the transcription factor dblGATA1, which exhibit a selective deficiency in eosinophils, demonstrated impaired A. fumigatus clearance and evidence of germinating organisms in the lung. Higher burden correlated with lower mRNA expression of Epx (eosinophil peroxidase) and Prg2 (major basic protein) as well as lower interleukin 1β (IL-1β), IL-6, IL-17A, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), and CXCL1 levels. However, examination of lung inflammatory cell populations failed to demonstrate defects in monocyte/macrophage, dendritic cell, or neutrophil recruitment in dblGATA1-deficient mice, suggesting that the absence of eosinophils in dlbGATA1-deficient mice was the sole cause of impaired lung clearance. We show that eosinophils generated from bone marrow have potent killing activity against A. fumigtaus in vitro, which does not require cell contact and can be recapitulated by eosinophil whole-cell lysates. Collectively, our data support a role for eosinophils in the lung response after A. fumigatus exposure.
Figures
Similar articles
-
Cytokine and chemokine responses following pulmonary challenge with Aspergillus fumigatus: obligatory role of TNF-alpha and GM-CSF in neutrophil recruitment.Med Mycol. 1999 Jun;37(3):183-94. doi: 10.1046/j.1365-280x.1999.00219.x. Med Mycol. 1999. PMID: 10421850
-
Dectin-1-dependent interleukin-22 contributes to early innate lung defense against Aspergillus fumigatus.Infect Immun. 2012 Jan;80(1):410-7. doi: 10.1128/IAI.05939-11. Epub 2011 Oct 28. Infect Immun. 2012. PMID: 22038916 Free PMC article.
-
Role of Granulocyte-Macrophage Colony-Stimulating Factor Signaling in Regulating Neutrophil Antifungal Activity and the Oxidative Burst During Respiratory Fungal Challenge.J Infect Dis. 2016 Apr 15;213(8):1289-98. doi: 10.1093/infdis/jiw054. Epub 2016 Feb 9. J Infect Dis. 2016. PMID: 26908736 Free PMC article.
-
Host defense mechanisms against Aspergillus fumigatus lung colonization and invasion.Curr Opin Microbiol. 2019 Dec;52:14-19. doi: 10.1016/j.mib.2019.04.003. Epub 2019 May 16. Curr Opin Microbiol. 2019. PMID: 31103956 Free PMC article. Review.
-
Innate Lung Defense during Invasive Aspergillosis: New Mechanisms.J Innate Immun. 2017;9(3):271-280. doi: 10.1159/000455125. Epub 2017 Feb 24. J Innate Immun. 2017. PMID: 28231567 Free PMC article. Review.
Cited by
-
Immune responses to invasive aspergillosis: new understanding and therapeutic opportunities.Curr Opin Infect Dis. 2017 Aug;30(4):364-371. doi: 10.1097/QCO.0000000000000381. Curr Opin Infect Dis. 2017. PMID: 28509673 Free PMC article. Review.
-
Factors Contributing to Sex Differences in Mice Inhaling Aspergillus fumigatus.Int J Environ Res Public Health. 2020 Nov 28;17(23):8851. doi: 10.3390/ijerph17238851. Int J Environ Res Public Health. 2020. PMID: 33260764 Free PMC article.
-
Evaluation of 2-[18F]-Fluorodeoxysorbitol PET Imaging in Preclinical Models of Aspergillus Infection.J Fungi (Basel). 2021 Dec 28;8(1):25. doi: 10.3390/jof8010025. J Fungi (Basel). 2021. PMID: 35049965 Free PMC article.
-
Eosinophils downregulate lung alloimmunity by decreasing TCR signal transduction.JCI Insight. 2019 Jun 6;4(11):e128241. doi: 10.1172/jci.insight.128241. eCollection 2019 Jun 6. JCI Insight. 2019. PMID: 31167966 Free PMC article.
-
Immune Interactions with Pathogenic and Commensal Fungi: A Two-Way Street.Immunity. 2015 Nov 17;43(5):845-58. doi: 10.1016/j.immuni.2015.10.023. Immunity. 2015. PMID: 26588778 Free PMC article. Review.
References
-
- Pappas PG, Alexander BD, Andes DR, Hadley S, Kauffman CA, Freifeld A, Anaissie EJ, Brumble LM, Herwaldt L, Ito J, Kontoyiannis DP, Lyon GM, Marr KA, Morrison VA, Park BJ, Patterson TF, Perl TM, Oster RA, Schuster MG, Walker R, Walsh TJ, Wannemuehler KA, Chiller TM. 2010. Invasive fungal infections among organ transplant recipients: results of the Transplant-Associated Infection Surveillance Network (TRANSNET). Clin. Infect. Dis. 50:1101–1111. 10.1086/651262 - DOI - PubMed
-
- Kontoyiannis DP, Marr KA, Park BJ, Alexander BD, Anaissie EJ, Walsh TJ, Ito J, Andes DR, Baddley JW, Brown JM, Brumble LM, Freifeld AG, Hadley S, Herwaldt LA, Kauffman CA, Knapp K, Lyon GM, Morrison VA, Papanicolaou G, Patterson TF, Perl TM, Schuster MG, Walker R, Wannemuehler KA, Wingard JR, Chiller TM, Pappas PG. 2010. Prospective surveillance for invasive fungal infections in hematopoietic stem cell transplant recipients, 2001–2006: overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) Database. Clin. Infect. Dis. 50:1091–1100. 10.1086/651263 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

