In vitro selection with artificial expanded genetic information systems
- PMID: 24379378
- PMCID: PMC3910645
- DOI: 10.1073/pnas.1311778111
In vitro selection with artificial expanded genetic information systems
Abstract
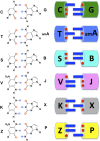
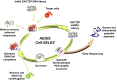
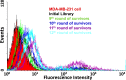
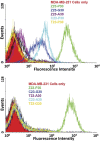
Artificially expanded genetic information systems (AEGISs) are unnatural forms of DNA that increase the number of independently replicating nucleotide building blocks. To do this, AEGIS pairs are joined by different arrangements of hydrogen bond donor and acceptor groups, all while retaining their Watson-Crick geometries. We report here a unique case where AEGIS DNA has been used to execute a systematic evolution of ligands by exponential enrichment (SELEX) experiment. This AEGIS-SELEX was designed to create AEGIS oligonucleotides that bind to a line of breast cancer cells. AEGIS-SELEX delivered an AEGIS aptamer (ZAP-2012) built from six different kinds of nucleotides (the standard G, A, C, and T, and the AEGIS nonstandard P and Z nucleotides, the last having a nitro functionality not found in standard DNA). ZAP-2012 has a dissociation constant of 30 nM against these cells. The affinity is diminished or lost when Z or P (or both) is replaced by standard nucleotides and compares well with affinities of standard GACT aptamers selected against cell lines using standard SELEX. The success of AEGIS-SELEX relies on various innovations, including (i) the ability to synthesize GACTZP libraries, (ii) polymerases that PCR amplify GACTZP DNA with little loss of the AEGIS nonstandard nucleotides, and (iii) technologies to deep sequence GACTZP DNA survivors. These results take the next step toward expanding the power and utility of SELEX and offer an AEGIS-SELEX that could possibly generate receptors, ligands, and catalysts having sequence diversities nearer to that displayed by proteins.
Keywords: cancer cell SELEX; next generation sequencing; nucleic acids; synthetic biology.
Conflict of interest statement
S.A.B., Z.Y., and the Foundation for Applied Molecular Evolution hold patents and patent applications for various of the AEGIS compounds and in vitro selection with expanded genetic alphabets.
Figures
References
-
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. - PubMed
-
- Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344(6265):467–468. - PubMed
-
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. - PubMed
-
- Famulok M. Oligonucleotide aptamers that recognize small molecules. Curr Opin Struct Biol. 1999;9(3):324–329. - PubMed
-
- Sun W, Du L, Li M. Aptamer-based carbohydrate recognition. Curr Pharm Des. 2010;16(20):2269–2278. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

