High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence
- PMID: 24379388
- PMCID: PMC3896158
- DOI: 10.1073/pnas.1315735111
High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence
Abstract
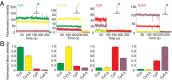
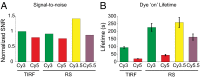
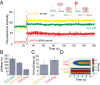
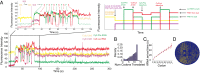
Zero-mode waveguides provide a powerful technology for studying single-molecule real-time dynamics of biological systems at physiological ligand concentrations. We customized a commercial zero-mode waveguide-based DNA sequencer for use as a versatile instrument for single-molecule fluorescence detection and showed that the system provides long fluorophore lifetimes with good signal to noise and low spectral cross-talk. We then used a ribosomal translation assay to show real-time fluidic delivery during data acquisition, showing it is possible to follow the conformation and composition of thousands of single biomolecules simultaneously through four spectral channels. This instrument allows high-throughput multiplexed dynamics of single-molecule biological processes over long timescales. The instrumentation presented here has broad applications to single-molecule studies of biological systems and is easily accessible to the biophysical community.
Conflict of interest statement
Conflict of interest statement: R.V.D., K.C., J. Cheng, M.E., P.-L.H., P.L., S.W.T., and D.R.H. are or were employees of Pacific Biosciences Inc., a company commercializing DNA sequencing technologies at the time that this work was completed.
Figures


References
-
- Moran-Mirabal JM, Craighead HG. Zero-mode waveguides: Sub-wavelength nanostructures for single molecule studies at high concentrations. Methods. 2008;46(1):11–17. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

