Fe-phyllosilicate redox cycling organisms from a redox transition zone in Hanford 300 Area sediments
- PMID: 24379809
- PMCID: PMC3863755
- DOI: 10.3389/fmicb.2013.00388
Fe-phyllosilicate redox cycling organisms from a redox transition zone in Hanford 300 Area sediments
Abstract
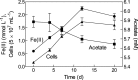
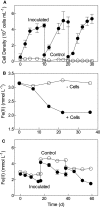
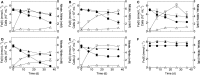
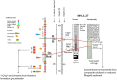
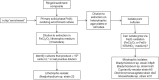
Microorganisms capable of reducing or oxidizing structural iron (Fe) in Fe-bearing phyllosilicate minerals were enriched and isolated from a subsurface redox transition zone at the Hanford 300 Area site in eastern Washington, USA. Both conventional and in situ "i-chip" enrichment strategies were employed. One Fe(III)-reducing Geobacter (G. bremensis strain R1, Deltaproteobacteria) and six Fe(II) phyllosilicate-oxidizing isolates from the Alphaproteobacteria (Bradyrhizobium japonicum strains 22, is5, and in8p8), Betaproteobacteria (Cupriavidus necator strain A5-1, Dechloromonas agitata strain is5), and Actinobacteria (Nocardioides sp. strain in31) were recovered. The G. bremensis isolate grew by oxidizing acetate with the oxidized form of NAu-2 smectite as the electron acceptor. The Fe(II)-oxidizers grew by oxidation of chemically reduced smectite as the energy source with nitrate as the electron acceptor. The Bradyrhizobium isolates could also carry out aerobic oxidation of biotite. This is the first report of the recovery of a Fe(II)-oxidizing Nocardioides, and to date only one other Fe(II)-oxidizing Bradyrhizobium is known. The 16S rRNA gene sequences of the isolates were similar to ones found in clone libraries from Hanford 300 sediments and groundwater, suggesting that such organisms may be present and active in situ. Whole genome sequencing of the isolates is underway, the results of which will enable comparative genomic analysis of mechanisms of extracellular phyllosilicate Fe redox metabolism, and facilitate development of techniques to detect the presence and expression of genes associated with microbial phyllosilicate Fe redox cycling in sediments.
Keywords: enrichment; iron; isolation; microbial; phyllosilicate; redox; sediment; subsurface.
Figures


Similar articles
-
Isolation of phyllosilicate-iron redox cycling microorganisms from an illite-smectite rich hydromorphic soil.Front Microbiol. 2012 Apr 4;3:134. doi: 10.3389/fmicb.2012.00134. eCollection 2012. Front Microbiol. 2012. PMID: 22493596 Free PMC article.
-
Microbial mineral colonization across a subsurface redox transition zone.Front Microbiol. 2015 Aug 28;6:858. doi: 10.3389/fmicb.2015.00858. eCollection 2015. Front Microbiol. 2015. PMID: 26379637 Free PMC article.
-
Growth and Population Dynamics of the Anaerobic Fe(II)-Oxidizing and Nitrate-Reducing Enrichment Culture KS.Appl Environ Microbiol. 2018 Apr 16;84(9):e02173-17. doi: 10.1128/AEM.02173-17. Print 2018 May 1. Appl Environ Microbiol. 2018. PMID: 29500257 Free PMC article.
-
Repeated anaerobic microbial redox cycling of iron.Appl Environ Microbiol. 2011 Sep;77(17):6036-42. doi: 10.1128/AEM.00276-11. Epub 2011 Jul 8. Appl Environ Microbiol. 2011. PMID: 21742920 Free PMC article.
-
Microbial iron-redox cycling in subsurface environments.Biochem Soc Trans. 2012 Dec 1;40(6):1249-56. doi: 10.1042/BST20120202. Biochem Soc Trans. 2012. PMID: 23176463 Review.
Cited by
-
Insights into Carbon Metabolism Provided by Fluorescence In Situ Hybridization-Secondary Ion Mass Spectrometry Imaging of an Autotrophic, Nitrate-Reducing, Fe(II)-Oxidizing Enrichment Culture.Appl Environ Microbiol. 2018 Apr 16;84(9):e02166-17. doi: 10.1128/AEM.02166-17. Print 2018 May 1. Appl Environ Microbiol. 2018. PMID: 29500258 Free PMC article.
-
Insights into Nitrate-Reducing Fe(II) Oxidation Mechanisms through Analysis of Cell-Mineral Associations, Cell Encrustation, and Mineralogy in the Chemolithoautotrophic Enrichment Culture KS.Appl Environ Microbiol. 2017 Jun 16;83(13):e00752-17. doi: 10.1128/AEM.00752-17. Print 2017 Jul 1. Appl Environ Microbiol. 2017. PMID: 28455336 Free PMC article.
-
Phylogenomic Analyses of Bradyrhizobium Reveal Uneven Distribution of the Lateral and Subpolar Flagellar Systems, Which Extends to Rhizobiales.Microorganisms. 2019 Feb 13;7(2):50. doi: 10.3390/microorganisms7020050. Microorganisms. 2019. PMID: 30781830 Free PMC article.
-
Phylogenetic distribution and evolutionary dynamics of nod and T3SS genes in the genus Bradyrhizobium.Microb Genom. 2020 Sep;6(9):mgen000407. doi: 10.1099/mgen.0.000407. Epub 2020 Aug 12. Microb Genom. 2020. PMID: 32783800 Free PMC article.
-
Hot in Cold: Microbial Life in the Hottest Springs in Permafrost.Microorganisms. 2020 Aug 27;8(9):1308. doi: 10.3390/microorganisms8091308. Microorganisms. 2020. PMID: 32867302 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

