Small proteins: untapped area of potential biological importance
- PMID: 24379829
- PMCID: PMC3864261
- DOI: 10.3389/fgene.2013.00286
Small proteins: untapped area of potential biological importance
Abstract
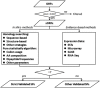
Polypeptides containing ≤100 amino acid residues (AAs) are generally considered to be small proteins (SPs). Many studies have shown that some SPs are involved in important biological processes, including cell signaling, metabolism, and growth. SP generally has a simple domain and has an advantage to be used as model system to overcome folding speed limits in protein folding simulation and drug design. But SPs were once thought to be trivial molecules in biological processes compared to large proteins. Because of the constraints of experimental methods and bioinformatics analysis, many genome projects have used a length threshold of 100 amino acid residues to minimize erroneous predictions and SPs are relatively under-represented in earlier studies. The general protein discovery methods have potential problems to predict and validate SPs, and very few effective tools and algorithms were developed specially for SPs identification. In this review, we mainly consider the diverse strategies applied to SPs prediction and discuss the challenge for differentiate SP coding genes from artifacts. We also summarize current large-scale discovery of SPs in species at the genome level. In addition, we present an overview of SPs with regard to biological significance, structural application, and evolution characterization in an effort to gain insight into the significance of SPs.
Keywords: evolution characterization; protein annotation coherence; protein identification; small ORFs; small proteins.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

