MicroRNA-132 is enriched in developing axons, locally regulates Rasa1 mRNA, and promotes axon extension
- PMID: 24381269
- PMCID: PMC3866495
- DOI: 10.1523/JNEUROSCI.3371-13.2014
MicroRNA-132 is enriched in developing axons, locally regulates Rasa1 mRNA, and promotes axon extension
Abstract
Developing axons can locally synthesize proteins, with roles in axon growth, guidance, and regeneration, but the mechanisms that regulate axonal mRNA translation are not well understood. MicroRNAs (miRNAs) are important regulators of translation but have still been little characterized in developing axons. Here we study mouse dorsal root ganglion (DRG) axons and show that their extension is impaired by conditional deficiency of the miRNA-processing enzyme Dicer in vitro and in vivo. A screen for axonal localization identifies a specific set of miRNAs preferentially enriched within the developing axon. High axonal expression and preferential localization were observed for miR-132, a miRNA previously known for roles in dendrites and dysregulation in major neurologic diseases. miR-132 knockdown reduced extension of cultured DRG axons, whereas overexpression increased extension. Mechanistically, miR-132 regulated the mRNA for the Ras GTPase activator Rasa1, a novel target in neuronal function. Moreover, miR-132 regulation of Rasa1 translation was seen in severed axons, demonstrating miRNA function locally within the axon. miR-132 expression in DRGs peaked in the period of maximum axon growth in vivo, consistent with its effect on axon growth, and suggesting a role as a developmental timer. Together, these findings identify miR-132 as a positive regulator of developing axon extension, acting through repression of Rasa1 mRNA, in a mechanism that operates locally within the axon.
Keywords: axon development; local mRNA translation; microRNA.
Figures
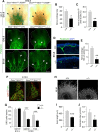


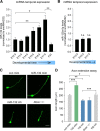

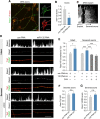
References
-
- Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, Scheppke L, Huang M, Shields DJ, Lindquist JN, Lapinski PE, King PD, Weis SM, Cheresh DA. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16:909–914. doi: 10.1038/nm.2186. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HD29417/HD/NICHD NIH HHS/United States
- EY01559/EY/NEI NIH HHS/United States
- R01 NS069913/NS/NINDS NIH HHS/United States
- RF1 NS069913/NS/NINDS NIH HHS/United States
- R37 HD029417/HD/NICHD NIH HHS/United States
- R56 NS069913/NS/NINDS NIH HHS/United States
- P30 HD018655/HD/NICHD NIH HHS/United States
- NS69913/NS/NINDS NIH HHS/United States
- R01 EY011559/EY/NEI NIH HHS/United States
- T32 NS007484/NS/NINDS NIH HHS/United States
- F32 NS073307/NS/NINDS NIH HHS/United States
- R01 HD029417/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
