Disruption of the ASTN2/TRIM32 locus at 9q33.1 is a risk factor in males for autism spectrum disorders, ADHD and other neurodevelopmental phenotypes
- PMID: 24381304
- PMCID: PMC3990173
- DOI: 10.1093/hmg/ddt669
Disruption of the ASTN2/TRIM32 locus at 9q33.1 is a risk factor in males for autism spectrum disorders, ADHD and other neurodevelopmental phenotypes
Abstract
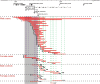
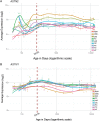
Rare copy number variants (CNVs) disrupting ASTN2 or both ASTN2 and TRIM32 have been reported at 9q33.1 by genome-wide studies in a few individuals with neurodevelopmental disorders (NDDs). The vertebrate-specific astrotactins, ASTN2 and its paralog ASTN1, have key roles in glial-guided neuronal migration during brain development. To determine the prevalence of astrotactin mutations and delineate their associated phenotypic spectrum, we screened ASTN2/TRIM32 and ASTN1 (1q25.2) for exonic CNVs in clinical microarray data from 89 985 individuals across 10 sites, including 64 114 NDD subjects. In this clinical dataset, we identified 46 deletions and 12 duplications affecting ASTN2. Deletions of ASTN1 were much rarer. Deletions near the 3' terminus of ASTN2, which would disrupt all transcript isoforms (a subset of these deletions also included TRIM32), were significantly enriched in the NDD subjects (P = 0.002) compared with 44 085 population-based controls. Frequent phenotypes observed in individuals with such deletions include autism spectrum disorder (ASD), attention deficit hyperactivity disorder (ADHD), speech delay, anxiety and obsessive compulsive disorder (OCD). The 3'-terminal ASTN2 deletions were significantly enriched compared with controls in males with NDDs, but not in females. Upon quantifying ASTN2 human brain RNA, we observed shorter isoforms expressed from an alternative transcription start site of recent evolutionary origin near the 3' end. Spatiotemporal expression profiling in the human brain revealed consistently high ASTN1 expression while ASTN2 expression peaked in the early embryonic neocortex and postnatal cerebellar cortex. Our findings shed new light on the role of the astrotactins in psychopathology and their interplay in human neurodevelopment.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- U01HG004446/HG/NHGRI NIH HHS/United States
- R01 DK058845/DK/NIDDK NIH HHS/United States
- P01 CA087969/CA/NCI NIH HHS/United States
- P01 CA055075/CA/NCI NIH HHS/United States
- P01 CA089392/CA/NCI NIH HHS/United States
- P50 DA019706/DA/NIDA NIH HHS/United States
- P50 CA084724/CA/NCI NIH HHS/United States
- CAPMC/ CIHR/Canada
- MH095867/MH/NIMH NIH HHS/United States
- U01 HG004399/HG/NHGRI NIH HHS/United States
- U01 HG004446/HG/NHGRI NIH HHS/United States
- U01HG004424/HG/NHGRI NIH HHS/United States
- CA87969/CA/NCI NIH HHS/United States
- DK58845/DK/NIDDK NIH HHS/United States
- CA55075/CA/NCI NIH HHS/United States
- U01 HG004424/HG/NHGRI NIH HHS/United States
- U01HG004399/HG/NHGRI NIH HHS/United States

