Guilt by rewiring: gene prioritization through network rewiring in genome wide association studies
- PMID: 24381306
- PMCID: PMC3990172
- DOI: 10.1093/hmg/ddt668
Guilt by rewiring: gene prioritization through network rewiring in genome wide association studies
Abstract
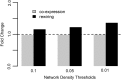
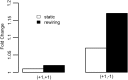
Although Genome Wide Association Studies (GWAS) have identified many susceptibility loci for common diseases, they only explain a small portion of heritability. It is challenging to identify the remaining disease loci because their association signals are likely weak and difficult to identify among millions of candidates. One potentially useful direction to increase statistical power is to incorporate functional genomics information, especially gene expression networks, to prioritize GWAS signals. Most current methods utilizing network information to prioritize disease genes are based on the 'guilt by association' principle, in which networks are treated as static, and disease-associated genes are assumed to locate closer with each other than random pairs in the network. In contrast, we propose a novel 'guilt by rewiring' principle. Studying the dynamics of gene networks between controls and patients, this principle assumes that disease genes more likely undergo rewiring in patients, whereas most of the network remains unaffected in disease condition. To demonstrate this principle, we consider the changes of co-expression networks in Crohn's disease patients and controls, and how network dynamics reveals information on disease associations. Our results demonstrate that network rewiring is abundant in the immune system, and disease-associated genes are more likely to be rewired in patients. To integrate this network rewiring feature and GWAS signals, we propose to use the Markov random field framework to integrate network information to prioritize genes. Applications in Crohn's disease and Parkinson's disease show that this framework leads to more replicable results, and implicates potentially disease-associated pathways.
Figures


References
-
- Manolio T.A., Brooks L.D., Collins F.S. A HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 2008;118:1590–1605. doi:10.1172/JCI34772. - DOI - PMC - PubMed
-
- Franke A., McGovern D.P.B., Barrett J.C., Wang K., Radford-Smith G.L., Ahmad T., Lees C.W., Balschun T., Lee J., Roberts R., et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi:10.1038/ng.717. - DOI - PMC - PubMed
-
- Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Philip Schumm L., Sharma Y., Anderson C.A., et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi:10.1038/nature11582. - DOI - PMC - PubMed
-
- Goldstein D.B. Common genetic variation and human traits. N. Engl. J. Med. 2009;360:1696–1698. doi:10.1056/NEJMp0806284. - DOI - PubMed
-
- Hardy J., Singleton A. Genomewide Association Studies and human disease. N. Engl. J. Med. 2009;360:1759–1768. doi:10.1056/NEJMra0808700. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

