Laboratory divergence of Methylobacterium extorquens AM1 through unintended domestication and past selection for antibiotic resistance
- PMID: 24384040
- PMCID: PMC3926354
- DOI: 10.1186/1471-2180-14-2
Laboratory divergence of Methylobacterium extorquens AM1 through unintended domestication and past selection for antibiotic resistance
Abstract
Background: A common assumption of microorganisms is that laboratory stocks will remain genetically and phenotypically constant over time, and across laboratories. It is becoming increasingly clear, however, that mutations can ruin strain integrity and drive the divergence or "domestication" of stocks. Since its discovery in 1960, a stock of Methylobacterium extorquens AM1 ("AM1") has remained in the lab, propagated across numerous growth and storage conditions, researchers, and facilities. To explore the extent to which this lineage has diverged, we compared our own "Modern" stock of AM1 to a sample archived at a culture stock center shortly after the strain's discovery. Stored as a lyophilized sample, we hypothesized that this Archival strain would better reflect the first-ever isolate of AM1 and reveal ways in which our Modern stock has changed through laboratory domestication or other means.
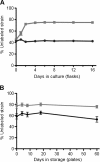
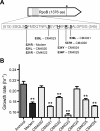
Results: Using whole-genome re-sequencing, we identified some 29 mutations - including single nucleotide polymorphisms, small indels, the insertion of mobile elements, and the loss of roughly 36 kb of DNA - that arose in the laboratory-maintained Modern lineage. Contrary to our expectations, Modern was both slower and less fit than Archival across a variety of growth substrates, and showed no improvement during long-term growth and storage. Modern did, however, outperform Archival during growth on nutrient broth, and in resistance to rifamycin, which was selected for by researchers in the 1980s. Recapitulating selection for rifamycin resistance in replicate Archival populations showed that mutations to RNA polymerase B (rpoB) substantially decrease growth in the absence of antibiotic, offering an explanation for slower growth in Modern stocks. Given the large number of genomic changes arising from domestication (28), it is somewhat surprising that the single other mutation attributed to purposeful laboratory selection accounts for much of the phenotypic divergence between strains.
Conclusions: These results highlight the surprising degree to which AM1 has diverged through a combination of unintended laboratory domestication and purposeful selection for rifamycin resistance. Instances of strain divergence are important, not only to ensure consistency of experimental results, but also to explore how microbes in the lab diverge from one another and from their wild counterparts.
Figures

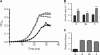

References
-
- Kearns DB, Losick R. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol. 2003;49:581–590. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

