Regulation of receptor tyrosine kinase ligand processing
- PMID: 24384567
- PMCID: PMC3941217
- DOI: 10.1101/cshperspect.a008995
Regulation of receptor tyrosine kinase ligand processing
Abstract
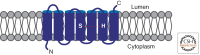
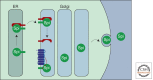
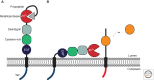
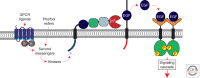
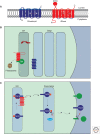
A primary mode of regulating receptor tyrosine kinase (RTK) signaling is to control access of ligand to its receptor. Many RTK ligands are synthesized as transmembrane proteins. Frequently, the active ligand must be released from the membrane by proteolysis before signaling can occur. Here, we discuss RTK ligand shedding and describe the proteases that catalyze it in flies and mammals. We focus principally on the control of EGF receptor ligand shedding, but also refer to ligands of other RTKs. Two prominent themes emerge. First, control by regulated trafficking and cellular compartmentalization of the proteases and their ligand substrates plays a key role in shedding. Second, many external signals converge on the shedding proteases and their control machinery. Proteases therefore act as regulatory hubs that integrate information that the cell receives and translate it into precise outgoing signals. The activation of signaling by proteases is therefore an essential element of the cellular communication machinery.
Figures
References
-
- Adrain C, Freeman M 2012. New lives for old: Evolution of pseudoenzyme function illustrated by iRhoms. Nat Rev Mol Cell Biol 13: 489–498 - PubMed
-
- Arduise C, Abache T, Li L, Billard M, Chabanon A, Ludwig A, Mauduit P, Boucheix C, Rubinstein E, Le Naour F 2008. Tetraspanins regulate ADAM10-mediated cleavage of TNF-α and epidermal growth factor. J Immunol 181: 7002–7013 - PubMed
-
- Asakura M, Kitakaze M, Takashima S, Liao Y, Ishikura F, Yoshinaka T, Ohmoto H, Node K, Yoshino K, Ishiguro H, et al. 2002. Cardiac hypertrophy is inhibited by antagonism of ADAM12 processing of HB-EGF: Metalloproteinase inhibitors as a new therapy. Nat Med 8: 35–40 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
