Gene model 129 (Gm129) encodes a novel transcriptional repressor that modulates circadian gene expression
- PMID: 24385426
- PMCID: PMC3931061
- DOI: 10.1074/jbc.M113.534651
Gene model 129 (Gm129) encodes a novel transcriptional repressor that modulates circadian gene expression
Abstract
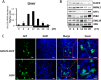
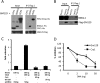
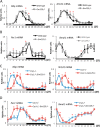
The mammalian circadian clock is a molecular oscillator composed of a feedback loop that involves transcriptional activators CLOCK and BMAL1, and repressors Cryptochrome (CRY) and Period (PER). Here we show that a direct CLOCK·BMAL1 target gene, Gm129, is a novel regulator of the feedback loop. ChIP analysis revealed that the CLOCK·BMAL1·CRY1 complex strongly occupies the promoter region of Gm129. Both mRNA and protein levels of GM129 exhibit high amplitude circadian oscillations in mouse liver, and Gm129 gene encodes a nuclear-localized protein that directly interacts with BMAL1 and represses CLOCK·BMAL1 activity. In vitro and in vivo protein-DNA interaction results demonstrate that, like CRY1, GM129 functions as a repressor by binding to the CLOCK·BMAL1 complex on DNA. Although Gm129(-/-) or Cry1(-/-) Gm129(-/-) mice retain a robust circadian rhythm, the peaks of Nr1d1 and Dbp mRNAs in liver exhibit a significant phase delay compared with control. Our results suggest that, in addition to CRYs and PERs, the GM129 protein contributes to the transcriptional feedback loop by modulating CLOCK·BMAL1 activity as a transcriptional repressor.
Keywords: Circadian; DNA-binding Protein; Gene Regulation; Signal Transduction; Transcription Repressor.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

