Local caspase activation interacts with Slit-Robo signaling to restrict axonal arborization
- PMID: 24385488
- PMCID: PMC3840933
- DOI: 10.1083/jcb.201303072
Local caspase activation interacts with Slit-Robo signaling to restrict axonal arborization
Abstract
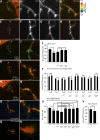
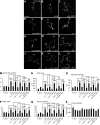
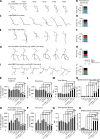
In addition to being critical for apoptosis, components of the apoptotic pathway, such as caspases, are involved in other physiological processes in many types of cells, including neurons. However, very little is known about their role in dynamic, nonphysically destructive processes, such as axonal arborization and synaptogenesis. We show that caspases were locally active in vivo at the branch points of young, dynamic retinal ganglion cell axonal arbors but not in the cell body or in stable mature arbors. Caspase activation, dependent on Caspase-3, Caspase-9, and p38 mitogen-activated protein kinase (MAPK), rapidly increased at branch points corresponding with branch tip addition. Time-lapse imaging revealed that knockdown of Caspase-3 and Caspase-9 led to more stable arbors and presynaptic sites. Genetic analysis showed that Caspase-3, Caspase-9, and p38 MAPK interacted with Slit1a-Robo2 signaling, suggesting that localized activation of caspases lie downstream of a ligand receptor system, acting as key promoters of axonal branch tip and synaptic dynamics to restrict arbor growth in vivo in the central nervous system.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

