Pfit is a structurally novel Crohn's disease-associated superantigen
- PMID: 24385909
- PMCID: PMC3873459
- DOI: 10.1371/journal.ppat.1003837
Pfit is a structurally novel Crohn's disease-associated superantigen
Abstract
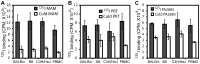
T cell responses to enteric bacteria are important in inflammatory bowel disease. I2, encoded by the pfiT gene of Pseudomonas fluorescens, is a T-cell superantigen associated with human Crohn's disease. Here we report the crystal structure of pfiT at 1.7Å resolution and provide a functional analysis of the interaction of pfiT and its homolog, PA2885, with human class II MHC. Both pfiT and PA2885 bound to mammalian cells and stimulated the proliferation of human lymphocytes. This binding was greatly inhibited by anti-class II MHC HLA-DR antibodies, and to a lesser extent, by anti HLA-DQ and DP antibodies, indicating that the binding was class II MHC-specific. GST-pfiT efficiently precipitated both endogenous and in vitro purified recombinant HLA-DR1 molecules, indicating that pfiT directly interacted with HLA-DR1. Competition studies revealed that pfiT and the superantigen Mycoplasma arthritidis mitogen (MAM) competed for binding to HLA-DR, indicating that their binding sites overlap. Structural analyses established that pfiT belongs to the TetR-family of DNA-binding transcription regulators. The distinct structure of pfiT indicates that it represents a new family of T cell superantigens.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
Pseudomonas fluorescens encodes the Crohn's disease-associated I2 sequence and T-cell superantigen.Infect Immun. 2002 Dec;70(12):6567-75. doi: 10.1128/IAI.70.12.6567-6575.2002. Infect Immun. 2002. PMID: 12438326 Free PMC article.
-
Zinc induces dimerization of the class II major histocompatibility complex molecule that leads to cooperative binding to a superantigen.J Biol Chem. 2007 Mar 2;282(9):5991-6000. doi: 10.1074/jbc.M608482200. Epub 2006 Dec 13. J Biol Chem. 2007. PMID: 17166841 Free PMC article.
-
A recombinant single-chain human class II MHC molecule (HLA-DR1) as a covalently linked heterotrimer of alpha chain, beta chain, and antigenic peptide, with immunogenicity in vitro and reduced affinity for bacterial superantigens.Eur J Immunol. 1997 Aug;27(8):1933-41. doi: 10.1002/eji.1830270817. Eur J Immunol. 1997. PMID: 9295029
-
Superantigens. Gazing into the crystal ball.Curr Biol. 1995 Mar 1;5(3):235-7. doi: 10.1016/s0960-9822(95)00047-9. Curr Biol. 1995. PMID: 7780728 Review.
-
Is celiac disease due to molecular mimicry between gliadin peptide-HLA class II molecule-T cell interactions and those of some unidentified superantigen?Mol Immunol. 1997 May;34(7):535-41. doi: 10.1016/s0161-5890(97)00068-0. Mol Immunol. 1997. PMID: 9364219 Review.
Cited by
-
Genomic and phenotypic insight into antimicrobial resistance of Pseudomonas fluorescens from King George Island, Antarctica.Front Microbiol. 2025 Mar 3;16:1535420. doi: 10.3389/fmicb.2025.1535420. eCollection 2025. Front Microbiol. 2025. PMID: 40099188 Free PMC article.
-
Autoimmunity and the microbiome: T-cell receptor mimicry of "self" and microbial antigens mediates self tolerance in holobionts: The concepts of "holoimmunity" (TcR-mediated tolerance for the holobiont) and "holoautoimmunity" (loss of tolerance for the holobiont) are introduced.Bioessays. 2016 Nov;38(11):1068-1083. doi: 10.1002/bies.201600083. Epub 2016 Sep 5. Bioessays. 2016. PMID: 27594308 Free PMC article.
-
Comparative genomics of Pseudomonas fluorescens subclade III strains from human lungs.BMC Genomics. 2015 Dec 7;16:1032. doi: 10.1186/s12864-015-2261-2. BMC Genomics. 2015. PMID: 26644001 Free PMC article.
References
-
- Kappler JW, Pullen A, Callahan J, Choi Y, Herman A, et al. (1989) Consequences of self and foreign superantigen interaction with specific V beta elements of the murine TCR alpha beta. Cold Spring Harb Symp Quant Biol 54 Pt 1: 401–407. - PubMed
-
- Marrack P, Kappler J (1990) The staphylococcal enterotoxins and their relatives. Science 248: 1066. - PubMed
-
- Kotzin BL, Leung DY, Kappler J, Marrack P (1993) Superantigens and their potential role in human disease. Adv Immunol 54: 99–166. - PubMed
-
- Li H, Llera A, Malchiodi EL, Mariuzza RA (1999) The structural basis of T cell activation by superantigens. Annu Rev Immunol 17: 435–466. - PubMed
-
- McCormick JK, Yarwood JM, Schlievert PM (2001) Toxic shock syndrome and bacterial superantigens: an update. Annu Rev Microbiol 55: 77–104. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

