Smc5/6 coordinates formation and resolution of joint molecules with chromosome morphology to ensure meiotic divisions
- PMID: 24385939
- PMCID: PMC3873251
- DOI: 10.1371/journal.pgen.1004071
Smc5/6 coordinates formation and resolution of joint molecules with chromosome morphology to ensure meiotic divisions
Abstract
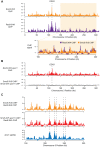
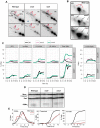
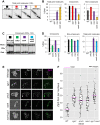
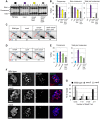
During meiosis, Structural Maintenance of Chromosome (SMC) complexes underpin two fundamental features of meiosis: homologous recombination and chromosome segregation. While meiotic functions of the cohesin and condensin complexes have been delineated, the role of the third SMC complex, Smc5/6, remains enigmatic. Here we identify specific, essential meiotic functions for the Smc5/6 complex in homologous recombination and the regulation of cohesin. We show that Smc5/6 is enriched at centromeres and cohesin-association sites where it regulates sister-chromatid cohesion and the timely removal of cohesin from chromosomal arms, respectively. Smc5/6 also localizes to recombination hotspots, where it promotes normal formation and resolution of a subset of joint-molecule intermediates. In this regard, Smc5/6 functions independently of the major crossover pathway defined by the MutLγ complex. Furthermore, we show that Smc5/6 is required for stable chromosomal localization of the XPF-family endonuclease, Mus81-Mms4(Eme1). Our data suggest that the Smc5/6 complex is required for specific recombination and chromosomal processes throughout meiosis and that in its absence, attempts at cell division with unresolved joint molecules and residual cohesin lead to severe recombination-induced meiotic catastrophe.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
-
- Kitajima TS, Kawashima SA, Watanabe Y (2004) The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427: 510–517. - PubMed
-
- Marston AL, Tham WH, Shah H, Amon A (2004) A genome-wide screen identifies genes required for centromeric cohesion. Science 303: 1367–1370. - PubMed
-
- Gregan J, Rabitsch PK, Sakem B, Csutak O, Latypov V, et al. (2005) Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr Biol 15: 1663–1669. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

