Loss of CLCA4 promotes epithelial-to-mesenchymal transition in breast cancer cells
- PMID: 24386311
- PMCID: PMC3873418
- DOI: 10.1371/journal.pone.0083943
Loss of CLCA4 promotes epithelial-to-mesenchymal transition in breast cancer cells
Abstract
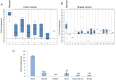
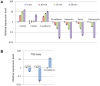
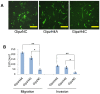
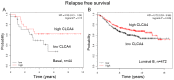
The epithelial to mesenchymal transition (EMT) is a developmental program in which epithelial cells downregulate their cell-cell junctions, acquire spindle cell morphology and exhibit cellular motility. In breast cancer, EMT facilitates invasion of surrounding tissues and correlates closely with cancer metastasis and relapse. We found previously that the candidate tumor suppressor CLCA2 is expressed in differentiated, growth-arrested mammary epithelial cells but is downregulated during tumor progression and EMT. We further demonstrated that CLCA2 is a p53-inducible proliferation-inhibitor whose loss indicates an increased risk of metastasis. We show here that another member of the CLCA gene family, CLCA4, is expressed in mammary epithelial cells and is similarly downregulated in breast tumors and in breast cancer cell lines. Like CLCA2, the gene is stress-inducible, and ectopic expression inhibits colony formation. Transcriptional profiling studies revealed that CLCA4 and CLCA2 together are markers for mammary epithelial differentiation, and both are downregulated by TGF beta. Moreover, knockdown of CLCA4 in immortalized cells by shRNAs caused downregulation of epithelial marker E-cadherin and CLCA2, while mesenchymal markers N-cadherin, vimentin, and fibronectin were upregulated. Double knockdown of CLCA2 and CLCA4 enhanced the mesenchymal profile. These findings suggest that CLCA4 and CLCA2 play complementary but distinct roles in epithelial differentiation. Clinically, low expression of CLCA4 signaled lower relapse-free survival in basal and luminal B breast cancers.
Conflict of interest statement
Figures




Similar articles
-
Calcium-Activated Chloride Channel A4 (CLCA4) Plays Inhibitory Roles in Invasion and Migration Through Suppressing Epithelial-Mesenchymal Transition via PI3K/AKT Signaling in Colorectal Cancer.Med Sci Monit. 2019 Jun 5;25:4176-4185. doi: 10.12659/MSM.914195. Med Sci Monit. 2019. PMID: 31164625 Free PMC article.
-
CLCA2 Interactor EVA1 Is Required for Mammary Epithelial Cell Differentiation.PLoS One. 2016 Mar 1;11(3):e0147489. doi: 10.1371/journal.pone.0147489. eCollection 2016. PLoS One. 2016. PMID: 26930581 Free PMC article.
-
CLCA4 inhibits cell proliferation and invasion of hepatocellular carcinoma by suppressing epithelial-mesenchymal transition via PI3K/AKT signaling.Aging (Albany NY). 2018 Oct 11;10(10):2570-2584. doi: 10.18632/aging.101571. Aging (Albany NY). 2018. PMID: 30312171 Free PMC article.
-
Along with its favorable prognostic role, CLCA2 inhibits growth and metastasis of nasopharyngeal carcinoma cells via inhibition of FAK/ERK signaling.J Exp Clin Cancer Res. 2018 Feb 20;37(1):34. doi: 10.1186/s13046-018-0692-8. J Exp Clin Cancer Res. 2018. PMID: 29463274 Free PMC article.
-
Epithelial protein lost in neoplasm (EPLIN): Beyond a tumor suppressor.Genes Dis. 2017 Apr 1;4(2):100-107. doi: 10.1016/j.gendis.2017.03.002. eCollection 2017 Jun. Genes Dis. 2017. PMID: 30258911 Free PMC article. Review.
Cited by
-
<em>miR-19a</em> targeting <em>CLCA4</em> to regulate the proliferation, migration, and invasion of colorectal cancer cells.Eur J Histochem. 2022 Mar 10;66(1):3381. doi: 10.4081/ejh.2022.3381. Eur J Histochem. 2022. PMID: 35266369 Free PMC article.
-
Interspecies diversity of chloride channel regulators, calcium-activated 3 genes.PLoS One. 2018 Jan 18;13(1):e0191512. doi: 10.1371/journal.pone.0191512. eCollection 2018. PLoS One. 2018. PMID: 29346439 Free PMC article.
-
Mutation profiling in eight cases of vagal paragangliomas.BMC Med Genomics. 2020 Sep 18;13(Suppl 8):115. doi: 10.1186/s12920-020-00763-4. BMC Med Genomics. 2020. PMID: 32948195 Free PMC article.
-
Calcium-Activated Chloride Channel A4 (CLCA4) Plays Inhibitory Roles in Invasion and Migration Through Suppressing Epithelial-Mesenchymal Transition via PI3K/AKT Signaling in Colorectal Cancer.Med Sci Monit. 2019 Jun 5;25:4176-4185. doi: 10.12659/MSM.914195. Med Sci Monit. 2019. PMID: 31164625 Free PMC article.
-
Proteomic analyses identify prognostic biomarkers for pancreatic ductal adenocarcinoma.Oncotarget. 2018 Jan 3;9(11):9789-9807. doi: 10.18632/oncotarget.23929. eCollection 2018 Feb 9. Oncotarget. 2018. PMID: 29515771 Free PMC article.
References
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. (2000) Molecular portraits of human breast tumours. Nature 406: 747–752. - PubMed
-
- Porter DA, Krop IE, Nasser S, Sgroi D, Kaelin CM, et al. (2001) A SAGE (serial analysis of gene expression) view of breast tumor progression. Cancer Res 61: 5697–5702. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

