Adaptation of an L-proline adenylation domain to use 4-propyl-L-proline in the evolution of lincosamide biosynthesis
- PMID: 24386435
- PMCID: PMC3874040
- DOI: 10.1371/journal.pone.0084902
Adaptation of an L-proline adenylation domain to use 4-propyl-L-proline in the evolution of lincosamide biosynthesis
Abstract
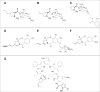
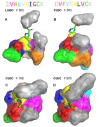
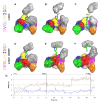
Clinically used lincosamide antibiotic lincomycin incorporates in its structure 4-propyl-L-proline (PPL), an unusual amino acid, while celesticetin, a less efficient related compound, makes use of proteinogenic L-proline. Biochemical characterization, as well as phylogenetic analysis and homology modelling combined with the molecular dynamics simulation were employed for complex comparative analysis of the orthologous protein pair LmbC and CcbC from the biosynthesis of lincomycin and celesticetin, respectively. The analysis proved the compared proteins to be the stand-alone adenylation domains strictly preferring their own natural substrate, PPL or L-proline. The LmbC substrate binding pocket is adapted to accommodate a rare PPL precursor. When compared with L-proline specific ones, several large amino acid residues were replaced by smaller ones opening a channel which allowed the alkyl side chain of PPL to be accommodated. One of the most important differences, that of the residue corresponding to V306 in CcbC changing to G308 in LmbC, was investigated in vitro and in silico. Moreover, the substrate binding pocket rearrangement also allowed LmbC to effectively adenylate 4-butyl-L-proline and 4-pentyl-L-proline, substrates with even longer alkyl side chains, producing more potent lincosamides. A shift of LmbC substrate specificity appears to be an integral part of biosynthetic pathway adaptation to the PPL acquisition. A set of genes presumably coding for the PPL biosynthesis is present in the lincomycin--but not in the celesticetin cluster; their homologs are found in biosynthetic clusters of some pyrrolobenzodiazepines (PBD) and hormaomycin. Whereas in the PBD and hormaomycin pathways the arising precursors are condensed to another amino acid moiety, the LmbC protein is the first functionally proved part of a unique condensation enzyme connecting PPL to the specialized amino sugar building unit.
Conflict of interest statement
Figures


References
-
- Brahme NM, Gonzalez JE, Rolls JP, Hessler EJ, Mizsak S et al. (1984) Biosynthesis of the lincomycins. 1. Studies using stable isotopes on the biosynthesis of the propyl-L-hygric and ethyl-L-hygric acid moieties of lincomycin-A and lincomycin-B. J Am Chem. Soc 106: 7873-7878.
-
- Neusser D, Schmidt H, Spizèk J, Novotnà J, Peschke U et al. (1998) The genes lmbB1 and lmbB2 of Streptomyces lincolnensis encode enzymes involved in the conversion of L-tyrosine to propylproline during the biosynthesis of the antibiotic lincomycin A. Arch Microbiol 169: 322-332. doi:10.1007/s002030050578. PubMed: 9531633. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

