Regulation of transcription factor activity by interconnected post-translational modifications
- PMID: 24388790
- PMCID: PMC3954851
- DOI: 10.1016/j.tips.2013.11.005
Regulation of transcription factor activity by interconnected post-translational modifications
Abstract
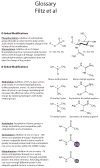
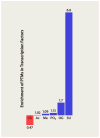
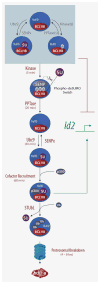
Transcription factors comprise just over 7% of the human proteome and serve as gatekeepers of cellular function, integrating external signal information into gene expression programs that reconfigure cellular physiology at the most basic levels. Surface-initiated cell signaling pathways converge on transcription factors, decorating these proteins with an array of post-translational modifications (PTMs) that are often interdependent, being linked in time, space, and combinatorial function. These PTMs orchestrate every activity of a transcription factor over its entire lifespan--from subcellular localization to protein-protein interactions, sequence-specific DNA binding, transcriptional regulatory activity, and protein stability--and play key roles in the epigenetic regulation of gene expression. The multitude of PTMs of transcription factors also offers numerous potential points of intervention for development of therapeutic agents to treat a wide spectrum of diseases. We review PTMs most commonly targeting transcription factors, focusing on recent reports of sequential and linked PTMs of individual factors.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures

References
-
- Holmberg CI, et al. Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem Sci. 2002;27:619–627. - PubMed
-
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. - PubMed
-
- Conaway RC, et al. Emerging roles of ubiquitin in transcription regulation. Science. 2002;296:1254–1258. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

