Structure of the human FANCL RING-Ube2T complex reveals determinants of cognate E3-E2 selection
- PMID: 24389026
- PMCID: PMC3979106
- DOI: 10.1016/j.str.2013.12.004
Structure of the human FANCL RING-Ube2T complex reveals determinants of cognate E3-E2 selection
Abstract
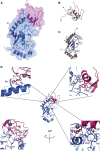
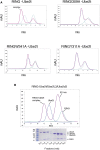
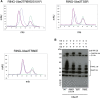
The combination of an E2 ubiquitin-conjugating enzyme with an E3 ubiquitin-ligase is essential for ubiquitin modification of a substrate. Moreover, the pairing dictates both the substrate choice and the modification type. The molecular details of generic E3-E2 interactions are well established. Nevertheless, the determinants of selective, specific E3-E2 recognition are not understood. There are ∼40 E2s and ∼600 E3s giving rise to a possible ∼24,000 E3-E2 pairs. Using the Fanconi Anemia pathway exclusive E3-E2 pair, FANCL-Ube2T, we report the atomic structure of the FANCL RING-Ube2T complex, revealing a specific and extensive network of additional electrostatic and hydrophobic interactions. Furthermore, we show that these specific interactions are required for selection of Ube2T over other E2s by FANCL.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Alpi A.F., Pace P.E., Babu M.M., Patel K.J. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol. Cell. 2008;32:767–777. - PubMed
-
- Alter B.P. Fanconi’s anemia and malignancies. Am. J. Hematol. 1996;53:99–110. - PubMed
-
- Bailly V., Lamb J., Sung P., Prakash S., Prakash L. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 1994;8:811–820. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

