Landscape of genomic alterations in cervical carcinomas
- PMID: 24390348
- PMCID: PMC4161954
- DOI: 10.1038/nature12881
Landscape of genomic alterations in cervical carcinomas
Abstract
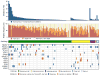
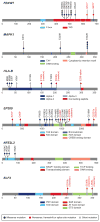
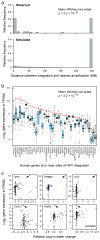
Cervical cancer is responsible for 10-15% of cancer-related deaths in women worldwide. The aetiological role of infection with high-risk human papilloma viruses (HPVs) in cervical carcinomas is well established. Previous studies have also implicated somatic mutations in PIK3CA, PTEN, TP53, STK11 and KRAS as well as several copy-number alterations in the pathogenesis of cervical carcinomas. Here we report whole-exome sequencing analysis of 115 cervical carcinoma-normal paired samples, transcriptome sequencing of 79 cases and whole-genome sequencing of 14 tumour-normal pairs. Previously unknown somatic mutations in 79 primary squamous cell carcinomas include recurrent E322K substitutions in the MAPK1 gene (8%), inactivating mutations in the HLA-B gene (9%), and mutations in EP300 (16%), FBXW7 (15%), NFE2L2 (4%), TP53 (5%) and ERBB2 (6%). We also observe somatic ELF3 (13%) and CBFB (8%) mutations in 24 adenocarcinomas. Squamous cell carcinomas have higher frequencies of somatic nucleotide substitutions occurring at cytosines preceded by thymines (Tp*C sites) than adenocarcinomas. Gene expression levels at HPV integration sites were statistically significantly higher in tumours with HPV integration compared with expression of the same genes in tumours without viral integration at the same site. These data demonstrate several recurrent genomic alterations in cervical carcinomas that suggest new strategies to combat this disease.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
-
- Jemal A, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. - PubMed
-
- IARC. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. 100B. International Agency for Research on Cancer; Lyon, France: 2012. A Review of Human Carcinogen: Biological Agents.
-
- zur Hausen H. Papillomaviruses in the causation of human cancers - a brief historical account. Virology. 2009;384:260–5. - PubMed
-
- Crook T, et al. Clonal p53 mutation in primary cervical cancer: association with human-papillomavirus-negative tumours. Lancet. 1992;339:1070–3. - PubMed
-
- McIntyre JB, et al. PIK3CA mutational status and overall survival in patients with cervical cancer treated with radical chemoradiotherapy. Gynecol Oncol. 2013;128:409–14. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

