Comparison and quantitative verification of mapping algorithms for whole-genome bisulfite sequencing
- PMID: 24391148
- PMCID: PMC3973287
- DOI: 10.1093/nar/gkt1325
Comparison and quantitative verification of mapping algorithms for whole-genome bisulfite sequencing
Abstract
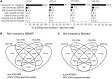
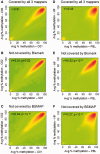
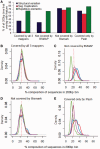
Coupling bisulfite conversion with next-generation sequencing (Bisulfite-seq) enables genome-wide measurement of DNA methylation, but poses unique challenges for mapping. However, despite a proliferation of Bisulfite-seq mapping tools, no systematic comparison of their genomic coverage and quantitative accuracy has been reported. We sequenced bisulfite-converted DNA from two tissues from each of two healthy human adults and systematically compared five widely used Bisulfite-seq mapping algorithms: Bismark, BSMAP, Pash, BatMeth and BS Seeker. We evaluated their computational speed and genomic coverage and verified their percentage methylation estimates. With the exception of BatMeth, all mappers covered >70% of CpG sites genome-wide and yielded highly concordant estimates of percentage methylation (r(2) ≥ 0.95). Fourfold variation in mapping time was found between BSMAP (fastest) and Pash (slowest). In each library, 8-12% of genomic regions covered by Bismark and Pash were not covered by BSMAP. An experiment using simulated reads confirmed that Pash has an exceptional ability to uniquely map reads in genomic regions of structural variation. Independent verification by bisulfite pyrosequencing generally confirmed the percentage methylation estimates by the mappers. Of these algorithms, Bismark provides an attractive combination of processing speed, genomic coverage and quantitative accuracy, whereas Pash offers considerably higher genomic coverage.
Figures


References
-
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

