Variant ionotropic receptors are expressed in olfactory sensory neurons of coeloconic sensilla on the antenna of the desert locust (Schistocerca gregaria)
- PMID: 24391446
- PMCID: PMC3879586
- DOI: 10.7150/ijbs.7624
Variant ionotropic receptors are expressed in olfactory sensory neurons of coeloconic sensilla on the antenna of the desert locust (Schistocerca gregaria)
Abstract
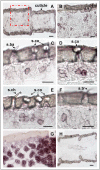
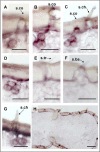
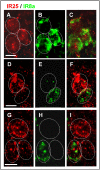
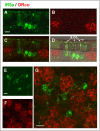
The behaviour of the desert locust, Schistocera gregaria, is largely directed by volatile olfactory cues. The relevant odorants are detected by specialized antennal sensory neurons which project their sensory dendrites into hair-like structures, the sensilla. Generally, the responsiveness of the antennal chemosensory cells is determined by specific receptors which may be either odorant receptors (ORs) or variant ionotropic receptors (IRs). Previously, we demonstrated that in locust the co-receptor for ORs (ORco) is only expressed in cells of sensilla basiconica and sensilla trichodea, suggesting that cells in sensilla coeloconica may express different types of chemosensory receptors. In this study, we have identified the genes of S. gregaria which encode homologues of co-receptors for the variant ionotropic receptors, the subtypes IR8a and IR25a. It was found that both subtypes, SgreIR8a and SgreIR25a, are expressed in the antennae of all five nymphal stages and in adults. Attempts to assign the relevant cell types by means of in situ hybridization revealed that SgreIR8a and SgreIR25a are expressed in cells of sensilla coeloconica. Double fluorescence in situ hybridization experiments disclosed that the two IR-subtypes are co-expressed in some cells of this sensillum type. Expression of SgreIR25a was also found in some of the sensilla chaetica, however, neither SgreIR25a nor SgreIR8a was found to be expressed in sensilla basiconica and sensilla trichodea. This observation was substantiated by the results of double FISH experiments demonstrating that cells expressing SgreIR8a or SgreIR25a do not express ORco. These results support the notion that the antenna of the desert locust employs two different populations of OSNs to sense odors; cells which express IRs in sensilla coeloconica and cells which express ORs in sensilla basiconica and sensilla trichodea.
Keywords: in situ hybridization; ionotropic receptors; locust; olfaction.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures





Similar articles
-
In Search for Pheromone Receptors: Certain Members of the Odorant Receptor Family in the Desert Locust Schistocerca gregaria (Orthoptera: Acrididae) Are Co-expressed with SNMP1.Int J Biol Sci. 2017 Jul 15;13(7):911-922. doi: 10.7150/ijbs.18402. eCollection 2017. Int J Biol Sci. 2017. PMID: 28808423 Free PMC article.
-
The olfactory co-receptor Orco from the migratory locust (Locusta migratoria) and the desert locust (Schistocerca gregaria): identification and expression pattern.Int J Biol Sci. 2012;8(2):159-70. doi: 10.7150/ijbs.8.159. Epub 2011 Dec 13. Int J Biol Sci. 2012. PMID: 22211114 Free PMC article.
-
Fine structure and distribution of antennal sensilla of the desert locust, Schistocerca gregaria (Orthoptera: Acrididae).Cell Tissue Res. 1998 Mar;291(3):525-36. doi: 10.1007/s004410051022. Cell Tissue Res. 1998. PMID: 9477309
-
Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond.Insect Biochem Mol Biol. 2013 Sep;43(9):888-97. doi: 10.1016/j.ibmb.2013.02.007. Epub 2013 Mar 1. Insect Biochem Mol Biol. 2013. PMID: 23459169 Review.
-
Elements of olfactory reception in adult Drosophila melanogaster.Anat Rec (Hoboken). 2013 Sep;296(9):1477-88. doi: 10.1002/ar.22747. Epub 2013 Jul 31. Anat Rec (Hoboken). 2013. PMID: 23904114 Review.
Cited by
-
The role of SNMPs in insect olfaction.Cell Tissue Res. 2021 Jan;383(1):21-33. doi: 10.1007/s00441-020-03336-0. Epub 2020 Nov 27. Cell Tissue Res. 2021. PMID: 33245414 Free PMC article. Review.
-
Ionotropic receptors in the turnip moth Agrotis segetum respond to repellent medium-chain fatty acids.BMC Biol. 2022 Feb 7;20(1):34. doi: 10.1186/s12915-022-01235-0. BMC Biol. 2022. PMID: 35130883 Free PMC article.
-
Identification and Characterization of Two "Sensory Neuron Membrane Proteins" (SNMPs) of the Desert Locust, Schistocerca gregaria (Orthoptera: Acrididae).J Insect Sci. 2016 Mar 24;16(1):33. doi: 10.1093/jisesa/iew015. Print 2016. J Insect Sci. 2016. PMID: 27012870 Free PMC article.
-
The antenna of horse stomach bot flies: morphology and phylogenetic implications (Oestridae, Gasterophilinae: Gasterophilus Leach).Sci Rep. 2016 Oct 5;6:34409. doi: 10.1038/srep34409. Sci Rep. 2016. PMID: 27703229 Free PMC article.
-
Haemocyte variations in 35 species of grasshoppers and locusts.Sci Prog. 2021 Oct;104(4):368504211053551. doi: 10.1177/00368504211053551. Sci Prog. 2021. PMID: 34751063 Free PMC article.
References
-
- Hassanali A, Njagi PG, Bashir MO. Chemical ecology of locusts and related acridids. Annu Rev Entomol. 2005;50:223–45. - PubMed
-
- Ochieng SA, Hallberg E, Hansson BS. Fine structure and distribution of antennal sensilla of the desert locust, Schistocerca gregaria (Orthoptera: Acrididae) Cell Tissue Res. 1998;291:525–36. - PubMed
-
- Ochieng SA, Hansson BS. Responses of olfactory receptor neurones to behaviourally important odours in gregarious and solitarious desert locust, Schistocerca gregaria. Physiol Entomol. 1999;24:28–36.
-
- Cui X, Wu C, Zhang L. Electrophysiological response patterns of 16 olfactory neurons from the trichoid sensilla to odorant from fecal volatiles in the locust, Locusta migratoria manilensis. Arch Insect Biochem Physiol. 2011;77:45–57. - PubMed
-
- Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373–91. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

