In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism
- PMID: 24391516
- PMCID: PMC3879213
- DOI: 10.1371/journal.pgen.1004047
In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism
Abstract
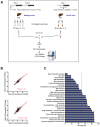
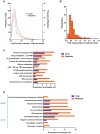
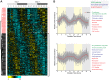
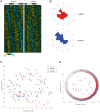
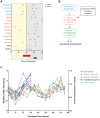
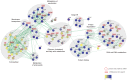
Circadian clocks are endogenous oscillators that drive the rhythmic expression of a broad array of genes, orchestrating metabolism and physiology. Recent evidence indicates that post-transcriptional and post-translational mechanisms play essential roles in modulating temporal gene expression for proper circadian function, particularly for the molecular mechanism of the clock. Due to technical limitations in large-scale, quantitative protein measurements, it remains unresolved to what extent the circadian clock regulates metabolism by driving rhythms of protein abundance. Therefore, we aimed to identify global circadian oscillations of the proteome in the mouse liver by applying in vivo SILAC mouse technology in combination with state of the art mass spectrometry. Among the 3000 proteins accurately quantified across two consecutive cycles, 6% showed circadian oscillations with a defined phase of expression. Interestingly, daily rhythms of one fifth of the liver proteins were not accompanied by changes at the transcript level. The oscillations of almost half of the cycling proteome were delayed by more than six hours with respect to the corresponding, rhythmic mRNA. Strikingly we observed that the length of the time lag between mRNA and protein cycles varies across the day. Our analysis revealed a high temporal coordination in the abundance of proteins involved in the same metabolic process, such as xenobiotic detoxification. Apart from liver specific metabolic pathways, we identified many other essential cellular processes in which protein levels are under circadian control, for instance vesicle trafficking and protein folding. Our large-scale proteomic analysis reveals thus that circadian post-transcriptional and post-translational mechanisms play a key role in the temporal orchestration of liver metabolism and physiology.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, et al. (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–320. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

