Comparative evolutionary and developmental dynamics of the cotton (Gossypium hirsutum) fiber transcriptome
- PMID: 24391525
- PMCID: PMC3879233
- DOI: 10.1371/journal.pgen.1004073
Comparative evolutionary and developmental dynamics of the cotton (Gossypium hirsutum) fiber transcriptome
Abstract
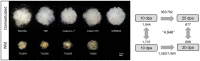
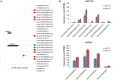
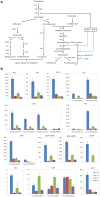
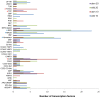
The single-celled cotton (Gossypium hirsutum) fiber provides an excellent model to investigate how human selection affects phenotypic evolution. To gain insight into the evolutionary genomics of cotton domestication, we conducted comparative transcriptome profiling of developing cotton fibers using RNA-Seq. Analysis of single-celled fiber transcriptomes from four wild and five domesticated accessions from two developmental time points revealed that at least one-third and likely one-half of the genes in the genome are expressed at any one stage during cotton fiber development. Among these, ~5,000 genes are differentially expressed during primary and secondary cell wall synthesis between wild and domesticated cottons, with a biased distribution among chromosomes. Transcriptome data implicate a number of biological processes affected by human selection, and suggest that the domestication process has prolonged the duration of fiber elongation in modern cultivated forms. Functional analysis suggested that wild cottons allocate greater resources to stress response pathways, while domestication led to reprogrammed resource allocation toward increased fiber growth, possibly through modulating stress-response networks. This first global transcriptomic analysis using multiple accessions of wild and domesticated cottons is an important step toward a more comprehensive systems perspective on cotton fiber evolution. The understanding that human selection over the past 5,000+ years has dramatically re-wired the cotton fiber transcriptome sets the stage for a deeper understanding of the genetic architecture underlying cotton fiber synthesis and phenotypic evolution.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Olsen KM, Wendel JF (2013) A bountiful harvest: genomic insights into crop domestication phenotypes. Annu Rev Plant Biol 64: 47–70. - PubMed
-
- Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127: 1309–1321. - PubMed
-
- Wendel JF, Flagel L, Adams KL (2012) Jeans, genes, and genomes: cotton as a model for studying polyploidy. In: Soltis PS, Soltis DE, editors. Polyploidy and Genome Evolution. Berlin: Springer. pp. 181–207.
-
- Wendel JF, Cronn RC (2003) Polyploidy and the evolutionary history of cotton. Adv Agron 78: 139–186.
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

