Comparison of metatranscriptomic samples based on k-tuple frequencies
- PMID: 24392128
- PMCID: PMC3879298
- DOI: 10.1371/journal.pone.0084348
Comparison of metatranscriptomic samples based on k-tuple frequencies
Abstract
Background: The comparison of samples, or beta diversity, is one of the essential problems in ecological studies. Next generation sequencing (NGS) technologies make it possible to obtain large amounts of metagenomic and metatranscriptomic short read sequences across many microbial communities. De novo assembly of the short reads can be especially challenging because the number of genomes and their sequences are generally unknown and the coverage of each genome can be very low, where the traditional alignment-based sequence comparison methods cannot be used. Alignment-free approaches based on k-tuple frequencies, on the other hand, have yielded promising results for the comparison of metagenomic samples. However, it is not known if these approaches can be used for the comparison of metatranscriptome datasets and which dissimilarity measures perform the best.
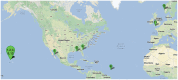
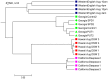
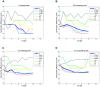
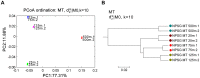
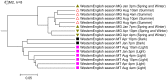
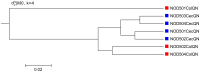
Results: We applied several beta diversity measures based on k-tuple frequencies to real metatranscriptomic datasets from pyrosequencing 454 and Illumina sequencing platforms to evaluate their effectiveness for the clustering of metatranscriptomic samples, including three d2-type dissimilarity measures, one dissimilarity measure in CVTree, one relative entropy based measure S2 and three classical 1p-norm distances. Results showed that the measure d2(S) can achieve superior performance on clustering metatranscriptomic samples into different groups under different sequencing depths for both 454 and Illumina datasets, recovering environmental gradients affecting microbial samples, classifying coexisting metagenomic and metatranscriptomic datasets, and being robust to sequencing errors. We also investigated the effects of tuple size and order of the background Markov model. A software pipeline to implement all the steps of analysis is built and is available at http://code.google.com/p/d2-tools/.
Conclusions: The k-tuple based sequence signature measures can effectively reveal major groups and gradient variation among metatranscriptomic samples from NGS reads. The d2(S) dissimilarity measure performs well in all application scenarios and its performance is robust with respect to tuple size and order of the Markov model.
Conflict of interest statement
Figures






References
-
- Smith T, Waterman M (1981) Comparison of biosequences. Adv Appl Math 2: 482–489.
-
- Altschul S, Gish W, Miller W, Myers E, Lipman Dea (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. - PubMed
-
- Dick GJ, Clement BG, Webb SM, Fodrie FJ, Bargar JR, et al. (2009) Enzymatic microbial Mn oxidation in the Guaymas Basin deep-sea hydrothermal plume. Geochim Cosmochim Ac 73: 6517–6530.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

