Ovarian cancer cell invasiveness is associated with discordant exosomal sequestration of Let-7 miRNA and miR-200
- PMID: 24393345
- PMCID: PMC3896684
- DOI: 10.1186/1479-5876-12-4
Ovarian cancer cell invasiveness is associated with discordant exosomal sequestration of Let-7 miRNA and miR-200
Abstract
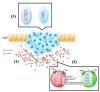
Background: The role of exosomes in the pathogenesis and metastatic spread of cancer remains to be fully elucidated. Recent studies support the hypothesis that the release of exosomes from cells modifies local extracellular conditions to promote cell growth and neovascularisation. In addition, exosomes may modify the phenotype of parent and/or target cell. For example, sequestration of signaling mediators into exosomes may reduce their intracellular bioavailability to the parent cell thereby altering cell phenotype and metastatic potential. The fusion of released exosomes with target cell and delivery may also modify cell function and activity. In this study, to further elucidate the role of exosomes in ovarian cancer, the release of exosomes from two ovarian cancer cell lines of different invasive capacity and their miRNA content of exosomes were compared. The hypothesis to be tested was that ovarian cancer cell invasiveness is associated with altered release of exosomes and discordant exosomal sequestration of miRNA.
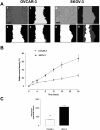
Methods: High (SKOV-3) and low (OVCAR-3) invasive ovarian cancer cell lines were used to characterize their exosome release. SKOV-3 and OVCAR-3 cells were cultured (DMEM, 20% exosome-free FBS) under an atmosphere of 8% O2 for 24 hours. Cell-conditioned media were collected and exosomes were isolated by differential and buoyant density centrifugation and characterised by Western blot (CD63 and CD9). Exosomal microRNA (let-7a-f and miR-200a-c) content was established by real-time PCR.
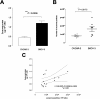
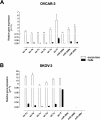
Results: Exosomes were identified with by the presence of typical cup-shaped spherical vesicle and the expression of exosome markers: CD63, CD9. SKOV-3 cells released 2.7-fold more exosomes (1.22 ± 0.11 μg/106 cells) compared to OVCAR-3 (0.44 ± 0.05 μg/106 cells). The let-7 family miRNA transcripts were identified in both ovarian cancer cell lines and their exosomes. The let-7 family transcripts were more abundant in OVCAR-3 cell than SKOV-3 cells. In contrast, let-7 family transcripts were more abundant in exosomes from SKOV-3 than OVCAR-3. miR-200 family transcripts were only identified in OVCAR-3 cells and their exosomes.
Conclusions: The data obtained in this study are consistent with the hypothesis that the releases of exosomes varies significantly between ovarian cancer cell lines and correlates with their invasive potential.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

