A novel intermediate in transcription initiation by human mitochondrial RNA polymerase
- PMID: 24393772
- PMCID: PMC3973326
- DOI: 10.1093/nar/gkt1356
A novel intermediate in transcription initiation by human mitochondrial RNA polymerase
Abstract
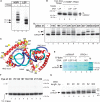
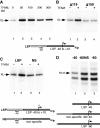
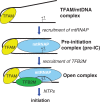
The mitochondrial genome is transcribed by a single-subunit T7 phage-like RNA polymerase (mtRNAP), structurally unrelated to cellular RNAPs. In higher eukaryotes, mtRNAP requires two transcription factors for efficient initiation-TFAM, a major nucleoid protein, and TFB2M, a transient component of mtRNAP catalytic site. The mechanisms behind assembly of the mitochondrial transcription machinery and its regulation are poorly understood. We isolated and identified a previously unknown human mitochondrial transcription intermediate-a pre-initiation complex that includes mtRNAP, TFAM and promoter DNA. Using protein-protein cross-linking, we demonstrate that human TFAM binds to the N-terminal domain of mtRNAP, which results in bending of the promoter DNA around mtRNAP. The subsequent recruitment of TFB2M induces promoter melting and formation of an open initiation complex. Our data indicate that the pre-initiation complex is likely to be an important target for transcription regulation and provide basis for further structural, biochemical and biophysical studies of mitochondrial transcription.
Figures



References
-
- Gaspari M, Larsson NG, Gustafsson CM. The transcription machinery in mammalian mitochondria. Biochim. Biophys. Acta. 2004;1659:148–152. - PubMed
-
- Ringel R, Sologub M, Morozov YI, Litonin D, Cramer P, Temiakov D. Structure of human mitochondrial RNA polymerase. Nature. 2011;478:269–273. - PubMed
-
- Gill SC, Yager TD, von Hippel PH. Thermodynamic analysis of the transcription cycle in E. coli. Biophys. Chem. 1990;37:239–250. - PubMed
-
- Buratowski S, Hahn S, Guarente L, Sharp PA. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. - PubMed
-
- Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

