The pseudogene TUSC2P promotes TUSC2 function by binding multiple microRNAs
- PMID: 24394498
- PMCID: PMC3896787
- DOI: 10.1038/ncomms3914
The pseudogene TUSC2P promotes TUSC2 function by binding multiple microRNAs
Abstract
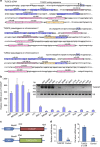
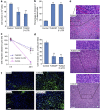
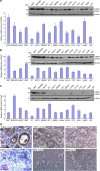
Various non-coding regions of the genome, once presumed to be 'junk' DNA, have recently been found to be transcriptionally active. In particular, pseudogenes are now known to have important biological roles. Here we report that transcripts of the two tumour suppressor candidate-2 pseudogenes (TUSC2P), found on chromosomes X and Y, are homologous to the 3'-UTR of their corresponding protein coding transcript, TUSC2. TUSC2P and the TUSC2 3'-UTR share many common miRNA-binding sites, including miR-17, miR-93, miR-299-3p, miR-520a, miR-608 and miR-661. We find that ectopic expression of TUSC2P and the TUSC2 3'-UTR inhibits cell proliferation, survival, migration, invasion and colony formation, and increases tumour cell death. By interacting with endogenous miRNAs, TUSC2P and TUSC2 3'-UTR arrest the functions of these miRNAs, resulting in increased translation of TUSC2. The TUSC2P and TUSC2 3'-UTR could thus be used as combinatorial miRNA inhibitors and might have clinical applications.
Figures






Similar articles
-
MicroRNA-663 facilitates the growth, migration and invasion of ovarian cancer cell by inhibiting TUSC2.Biol Res. 2019 Apr 3;52(1):18. doi: 10.1186/s40659-019-0219-6. Biol Res. 2019. PMID: 30944041 Free PMC article.
-
TUSC2P suppresses the tumor function of esophageal squamous cell carcinoma by regulating TUSC2 expression and correlates with disease prognosis.BMC Cancer. 2018 Sep 15;18(1):894. doi: 10.1186/s12885-018-4804-9. BMC Cancer. 2018. PMID: 30219035 Free PMC article.
-
miR-632 Promotes Laryngeal Carcinoma Cell Proliferation, Migration, and Invasion Through Negative Regulation of GSK3β.Oncol Res. 2020 Feb 7;28(1):21-31. doi: 10.3727/096504018X15213142076069. Epub 2018 Mar 21. Oncol Res. 2020. PMID: 29562960 Free PMC article.
-
MicroRNA-138 is a Prognostic Biomarker for Triple-Negative Breast Cancer and Promotes Tumorigenesis via TUSC2 repression.Sci Rep. 2019 Sep 3;9(1):12718. doi: 10.1038/s41598-019-49155-4. Sci Rep. 2019. PMID: 31481748 Free PMC article.
-
Tumor Suppressor Candidate 2 (TUSC2): Discovery, Functions, and Cancer Therapy.Cancers (Basel). 2023 Apr 25;15(9):2455. doi: 10.3390/cancers15092455. Cancers (Basel). 2023. PMID: 37173921 Free PMC article. Review.
Cited by
-
TWIST1/miR-584/TUSC2 pathway induces resistance to apoptosis in thyroid cancer cells.Oncotarget. 2016 Oct 25;7(43):70575-70588. doi: 10.18632/oncotarget.12129. Oncotarget. 2016. PMID: 27661106 Free PMC article.
-
MicroRNA-663 facilitates the growth, migration and invasion of ovarian cancer cell by inhibiting TUSC2.Biol Res. 2019 Apr 3;52(1):18. doi: 10.1186/s40659-019-0219-6. Biol Res. 2019. PMID: 30944041 Free PMC article.
-
A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy.Oncogene. 2018 Nov;37(44):5829-5842. doi: 10.1038/s41388-018-0369-y. Epub 2018 Jul 4. Oncogene. 2018. PMID: 29973691
-
Tumor suppressor candidate 2 (TUSC2, FUS-1) and human cancers.Discov Med. 2017 May;23(128):325-330. Discov Med. 2017. PMID: 28715648 Free PMC article. Review.
-
MiR-608, pre-miR-124-1 and pre-miR26a-1 polymorphisms modify susceptibility and recurrence-free survival in surgically resected CRC individuals.Oncotarget. 2016 Nov 15;7(46):75865-75873. doi: 10.18632/oncotarget.12422. Oncotarget. 2016. PMID: 27713147 Free PMC article.
References
-
- Hutvagner G. & Zamore P. D. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297, 2056–2060 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

