microRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial-mesenchymal transition
- PMID: 24394555
- PMCID: PMC3896774
- DOI: 10.1038/ncomms3977
microRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial-mesenchymal transition
Abstract
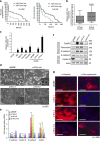
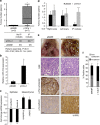
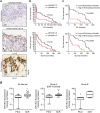
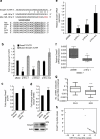
Ovarian cancer is a leading cause of cancer deaths among women. Effective targets to treat advanced epithelial ovarian cancer (EOC) and biomarkers to predict treatment response are still lacking because of the complexity of pathways involved in ovarian cancer progression. Here we show that miR-181a promotes TGF-β-mediated epithelial-to-mesenchymal transition via repression of its functional target, Smad7. miR-181a and phosphorylated Smad2 are enriched in recurrent compared with matched-primary ovarian tumours and their expression is associated with shorter time to recurrence and poor outcome in patients with EOC. Furthermore, ectopic expression of miR-181a results in increased cellular survival, migration, invasion, drug resistance and in vivo tumour burden and dissemination. In contrast, miR-181a inhibition via decoy vector suppression and Smad7 re-expression results in significant reversion of these phenotypes. Combined, our findings highlight an unappreciated role for miR-181a, Smad7, and the TGF-β signalling pathway in high-grade serous ovarian cancer.
Figures




Similar articles
-
The activation of microRNA-520h-associated TGF-β1/c-Myb/Smad7 axis promotes epithelial ovarian cancer progression.Cell Death Dis. 2018 Aug 29;9(9):884. doi: 10.1038/s41419-018-0946-6. Cell Death Dis. 2018. PMID: 30158641 Free PMC article.
-
Hepatocyte growth factor secreted by ovarian cancer cells stimulates peritoneal implantation via the mesothelial-mesenchymal transition of the peritoneum.Gynecol Oncol. 2015 Nov;139(2):345-54. doi: 10.1016/j.ygyno.2015.08.010. Epub 2015 Aug 31. Gynecol Oncol. 2015. PMID: 26335595
-
Low MAD2 expression levels associate with reduced progression-free survival in patients with high-grade serous epithelial ovarian cancer.J Pathol. 2012 Apr;226(5):746-55. doi: 10.1002/path.3035. Epub 2012 Jan 17. J Pathol. 2012. PMID: 22069160 Free PMC article.
-
Ovarian serous carcinogenesis from tubal secretory cells.Histol Histopathol. 2015 Nov;30(11):1295-302. doi: 10.14670/HH-11-645. Epub 2015 Jul 15. Histol Histopathol. 2015. PMID: 26174492 Review.
-
Targeting pathways contributing to epithelial-mesenchymal transition (EMT) in epithelial ovarian cancer.Curr Drug Targets. 2012 Dec;13(13):1649-53. doi: 10.2174/138945012803530044. Curr Drug Targets. 2012. PMID: 23061545 Review.
Cited by
-
Metformin inhibits tumor growth by regulating multiple miRNAs in human cholangiocarcinoma.Oncotarget. 2015 Feb 20;6(5):3178-94. doi: 10.18632/oncotarget.3063. Oncotarget. 2015. PMID: 25605008 Free PMC article.
-
Quercetin up-regulates the expression of tumor-suppressive microRNAs in human cervical cancer.Biosci Microbiota Food Health. 2023;42(1):87-93. doi: 10.12938/bmfh.2022-056. Epub 2022 Oct 5. Biosci Microbiota Food Health. 2023. PMID: 36660602 Free PMC article.
-
Role of miR‑181a‑5p in cancer (Review).Int J Oncol. 2023 Oct;63(4):108. doi: 10.3892/ijo.2023.5556. Epub 2023 Aug 4. Int J Oncol. 2023. PMID: 37539738 Free PMC article. Review.
-
MicroRNA: a new and promising potential biomarker for diagnosis and prognosis of ovarian cancer.Cancer Biol Med. 2015 Dec;12(4):328-41. doi: 10.7497/j.issn.2095-3941.2015.0024. Cancer Biol Med. 2015. PMID: 26779370 Free PMC article. Review.
-
Epigallocatechin-3-O-gallate up-regulates microRNA-let-7b expression by activating 67-kDa laminin receptor signaling in melanoma cells.Sci Rep. 2016 Jan 12;6:19225. doi: 10.1038/srep19225. Sci Rep. 2016. PMID: 26754091 Free PMC article.
References
-
- Siegel R., Ward E., Brawley O. & Jemal A. Cancer statistics. Cancer J. Clin. 61, 212–216 (2011). - PubMed
-
- Seidman J. D. et al. Borderline ovarian tumors: diverse contemporary viewpoints on terminology and diagnostic criteria with illustrative images. Hum. Pathol. 35, 918–933 (2004). - PubMed
-
- Kumar M. S., Lu J., Mercer K. L., Golub T. R. & Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 39, 673–677 (2007). - PubMed
-
- Mateescu B. et al. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat. Med. 17, 1627–1635 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

