CLOCK:BMAL1 is a pioneer-like transcription factor
- PMID: 24395244
- PMCID: PMC3894415
- DOI: 10.1101/gad.228536.113
CLOCK:BMAL1 is a pioneer-like transcription factor
Abstract
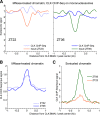
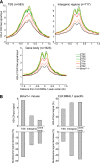
The mammalian circadian clock relies on the master genes CLOCK and BMAL1 to drive rhythmic gene expression and regulate biological functions under circadian control. Here we show that rhythmic CLOCK:BMAL1 DNA binding promotes rhythmic chromatin opening. Mechanisms include CLOCK:BMAL1 binding to nucleosomes and rhythmic chromatin modification; e.g., incorporation of the histone variant H2A.Z. This rhythmic chromatin remodeling mediates the rhythmic binding of other transcription factors adjacent to CLOCK:BMAL1, suggesting that the activity of these other transcription factors contributes to the genome-wide CLOCK:BMAL1 heterogeneous transcriptional output. These data therefore indicate that the clock regulation of transcription relies on the rhythmic regulation of chromatin accessibility and suggest that the concept of pioneer function extends to acute gene regulation.
Keywords: H2A.Z; MNase-seq; chromatin modifications; circadian rhythms; nucleosome occupancy; nucleosome positioning; regulation of transcription.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
