Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization
- PMID: 24395781
- PMCID: PMC3903257
- DOI: 10.1073/pnas.1312099111
Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization
Abstract
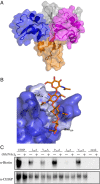
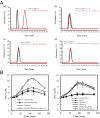
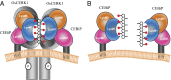
Perception of microbe-associated molecular patterns (MAMPs) through pattern recognition receptors (PRRs) triggers various defense responses in plants. This MAMP-triggered immunity plays a major role in the plant resistance against various pathogens. To clarify the molecular basis of the specific recognition of chitin oligosaccharides by the rice PRR, CEBiP (chitin-elicitor binding protein), as well as the formation and activation of the receptor complex, biochemical, NMR spectroscopic, and computational studies were performed. Deletion and domain-swapping experiments showed that the central lysine motif in the ectodomain of CEBiP is essential for the binding of chitin oligosaccharides. Epitope mapping by NMR spectroscopy indicated the preferential binding of longer-chain chitin oligosaccharides, such as heptamer-octamer, to CEBiP, and also the importance of N-acetyl groups for the binding. Molecular modeling/docking studies clarified the molecular interaction between CEBiP and chitin oligosaccharides and indicated the importance of Ile122 in the central lysine motif region for ligand binding, a notion supported by site-directed mutagenesis. Based on these results, it was indicated that two CEBiP molecules simultaneously bind to one chitin oligosaccharide from the opposite side, resulting in the dimerization of CEBiP. The model was further supported by the observations that the addition of (GlcNAc)8 induced dimerization of the ectodomain of CEBiP in vitro, and the dimerization and (GlcNAc)8-induced reactive oxygen generation were also inhibited by a unique oligosaccharide, (GlcNβ1,4GlcNAc)4, which is supposed to have N-acetyl groups only on one side of the molecule. Based on these observations, we proposed a hypothetical model for the ligand-induced activation of a receptor complex, involving both CEBiP and Oryza sativa chitin-elicitor receptor kinase-1.
Keywords: LysM-receptor; MTI/PTI; chitin signaling; plant immunity; receptor–ligand interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Zipfel C. Pattern-recognition receptors in plant innate immunity. Curr Opin Immunol. 2008;20(1):10–16. - PubMed
-
- Kwon C, Panstruga R, Schulze-Lefert P. Les liaisons dangereuses: Immunological synapse formation in animals and plants. Trends Immunol. 2008;29(4):159–166. - PubMed
-
- Nürnberger T, Brunner F, Kemmerling B, Piater L. Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol Rev. 2004;198:249–266. - PubMed
-
- Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

