X13CMS: global tracking of isotopic labels in untargeted metabolomics
- PMID: 24397582
- PMCID: PMC3982964
- DOI: 10.1021/ac403384n
X13CMS: global tracking of isotopic labels in untargeted metabolomics
Abstract
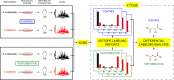
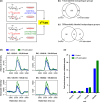
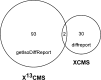
Studies of isotopically labeled compounds have been fundamental to understanding metabolic pathways and fluxes. They have traditionally, however, been used in conjunction with targeted analyses that identify and quantify a limited number of labeled downstream metabolites. Here we describe an alternative workflow that leverages recent advances in untargeted metabolomic technologies to track the fates of isotopically labeled metabolites in a global, unbiased manner. This untargeted approach can be applied to discover novel biochemical pathways and characterize changes in the fates of labeled metabolites as a function of altered biological conditions such as disease. To facilitate the data analysis, we introduce X(13)CMS, an extension of the widely used mass spectrometry-based metabolomic software package XCMS. X(13)CMS uses the XCMS platform to detect metabolite peaks and perform retention-time alignment in liquid chromatography/mass spectrometry (LC/MS) data. With the use of the XCMS output, the program then identifies isotopologue groups that correspond to isotopically labeled compounds. The retrieval of these groups is done without any a priori knowledge besides the following input parameters: (i) the mass difference between the unlabeled and labeled isotopes, (ii) the mass accuracy of the instrument used in the analysis, and (iii) the estimated retention-time reproducibility of the chromatographic method. Despite its name, X(13)CMS can be used to track any isotopic label. Additionally, it detects differential labeling patterns in biological samples collected from parallel control and experimental conditions. We validated the ability of X(13)CMS to accurately retrieve labeled metabolites from complex biological matrices both with targeted LC/MS/MS analysis of a subset of the hits identified by the program and with labeled standards spiked into cell extracts. We demonstrate the full functionality of X(13)CMS with an analysis of cultured rat astrocytes treated with uniformly labeled (U-)(13)C-glucose during lipopolysaccharide (LPS) challenge. Our results show that out of 223 isotopologue groups enriched from U-(13)C-glucose, 95 have statistically significant differential labeling patterns in astrocytes challenged with LPS compared to unchallenged control cells. Only two of these groups overlap with the 32 differentially regulated peaks identified by XCMS, indicating that X(13)CMS uncovers different and complementary information from untargeted metabolomic studies. Like XCMS, X(13)CMS is implemented in R. It is available from our laboratory website at http://pattilab.wustl.edu/x13cms.php .
Figures

References
-
- Schoenheimer R.; Rittenberg D. Science 1938, 87, 221–226. - PubMed
-
- Wolfe R. R.; Chinkes D. L.. Isotope Tracers in Metabolic Research: Principles and Practice of Kinetic Analysis, 2nd ed.; Wiley-Liss: Hoboken, NJ, 2005.
-
- Bloch K.; Rittenberg D. J. Biol. Chem. 1942, 145, 625.
-
- Bloch K.; Rittenberg D. J. Biol. Chem. 1945, 159, 45. - PubMed
-
- Entner N.; Doudoroff M. J. Biol. Chem. 1952, 196, 853–862. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

