Nuclear export signal within CALM is necessary for CALM-AF10-induced leukemia
- PMID: 24397609
- PMCID: PMC4317939
- DOI: 10.1111/cas.12347
Nuclear export signal within CALM is necessary for CALM-AF10-induced leukemia
Abstract
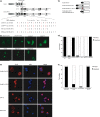
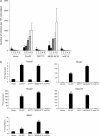
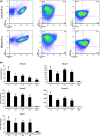
The CALM-AF10 fusion gene, which results from a t(10;11) translocation, is found in a variety of hematopoietic malignancies. Certain HOXA cluster genes and MEIS1 genes are upregulated in patients and mouse models that express CALM-AF10. Wild-type clathrin assembly lymphoid myeloid leukemia protein (CALM) primarily localizes in a diffuse pattern within the cytoplasm, whereas AF10 localizes in the nucleus; however, it is not clear where CALM-AF10 acts to induce leukemia. To investigate the influence of localization on leukemogenesis involving CALM-AF10, we determined the nuclear export signal (NES) within CALM that is necessary and sufficient for cytoplasmic localization of CALM-AF10. Mutations in the NES eliminated the capacity of CALM-AF10 to immortalize murine bone-marrow cells in vitro and to promote development of acute myeloid leukemia in mouse models. Furthermore, a fusion of AF10 with the minimal NES can immortalize bone-marrow cells and induce leukemia in mice. These results suggest that during leukemogenesis, CALM-AF10 plays its critical roles in the cytoplasm.
Keywords: AF10; chromosome translocation; histone modification; leukemia; nuclear export signal.
© 2014 The Authors. Cancer Science published by Wiley Publishing Asia Pty Ltd on behalf of Japanese Cancer Association.
Figures


Similar articles
-
A CALM-derived nuclear export signal is essential for CALM-AF10-mediated leukemogenesis.Blood. 2013 Jun 6;121(23):4758-68. doi: 10.1182/blood-2012-06-435792. Epub 2013 Mar 13. Blood. 2013. PMID: 23487024 Free PMC article.
-
The leukemogenic CALM/AF10 fusion protein alters the subcellular localization of the lymphoid regulator Ikaros.Oncogene. 2008 May 1;27(20):2886-96. doi: 10.1038/sj.onc.1210945. Epub 2007 Nov 26. Oncogene. 2008. PMID: 18037964
-
Leukaemic transformation by CALM-AF10 involves upregulation of Hoxa5 by hDOT1L.Nat Cell Biol. 2006 Sep;8(9):1017-24. doi: 10.1038/ncb1464. Epub 2006 Aug 20. Nat Cell Biol. 2006. PMID: 16921363 Free PMC article.
-
Clathrin assembly lymphoid myeloid leukemia-AF10-positive acute leukemias: a report of 2 cases with a review of the literature.Korean J Lab Med. 2010 Apr;30(2):117-21. doi: 10.3343/kjlm.2010.30.2.117. Korean J Lab Med. 2010. PMID: 20445327 Review.
-
The role of CALM-AF10 gene fusion in acute leukemia.Leukemia. 2008 Apr;22(4):678-85. doi: 10.1038/sj.leu.2405074. Epub 2007 Dec 20. Leukemia. 2008. PMID: 18094714 Free PMC article. Review.
Cited by
-
The role of the PZP domain of AF10 in acute leukemia driven by AF10 translocations.Nat Commun. 2021 Jul 5;12(1):4130. doi: 10.1038/s41467-021-24418-9. Nat Commun. 2021. PMID: 34226546 Free PMC article.
-
MOZ/ENL complex is a recruiting factor of leukemic AF10 fusion proteins.Nat Commun. 2023 Apr 8;14(1):1979. doi: 10.1038/s41467-023-37712-5. Nat Commun. 2023. PMID: 37031220 Free PMC article.
-
Prognostic relevance of genetic variations in T-cell acute lymphoblastic leukemia/lymphoblastic lymphoma.Transl Cancer Res. 2019 Oct;8(6):2485-2495. doi: 10.21037/tcr.2019.10.04. Transl Cancer Res. 2019. PMID: 35117001 Free PMC article. Review.
-
The Biochemical Properties and Functions of CALM and AP180 in Clathrin Mediated Endocytosis.Membranes (Basel). 2014 Jul 31;4(3):388-413. doi: 10.3390/membranes4030388. Membranes (Basel). 2014. PMID: 25090048 Free PMC article.
-
The ∼ 16 kDa C-terminal sequence of clathrin assembly protein AP180 is essential for efficient clathrin binding.PLoS One. 2014 Oct 20;9(10):e110557. doi: 10.1371/journal.pone.0110557. eCollection 2014. PLoS One. 2014. PMID: 25329427 Free PMC article.
References
-
- Bohlander SK, Muschinsky V, Schrader K, et al. Molecular analysis of the CALM/AF10 fusion: identical rearrangements in acute myeloid leukemia, acute lymphoblastic leukemia and malignant lymphoma patients. Leukemia. 2000;14:93–9. - PubMed
-
- Narita M, Shimizu K, Hayashi Y, et al. Consistent detection of CALM-AF10 chimaeric transcripts in haematological malignancies with t(10;11)(p13;q14) and identification of novel transcripts. Br J Haematol. 1999;105:928–37. - PubMed
-
- Linder B, Newman R, Jones LK, et al. Biochemical analyses of the AF10 protein: the extended LAP/PHD-finger mediates oligomerisation. J Mol Biol. 2000;299:369–78. - PubMed
-
- Wysocka J, Swigut T, Xiao H, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

