Phylogenomic test of the hypotheses for the evolutionary origin of eukaryotes
- PMID: 24398320
- PMCID: PMC3969559
- DOI: 10.1093/molbev/mst272
Phylogenomic test of the hypotheses for the evolutionary origin of eukaryotes
Abstract
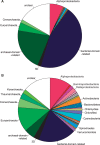
The evolutionary origin of eukaryotes is a question of great interest for which many different hypotheses have been proposed. These hypotheses predict distinct patterns of evolutionary relationships for individual genes of the ancestral eukaryotic genome. The availability of numerous completely sequenced genomes covering the three domains of life makes it possible to contrast these predictions with empirical data. We performed a systematic analysis of the phylogenetic relationships of ancestral eukaryotic genes with archaeal and bacterial genes. In contrast with previous studies, we emphasize the critical importance of methods accounting for statistical support, horizontal gene transfer, and gene loss, and we disentangle the processes underlying the phylogenomic pattern we observe. We first recover a clear signal indicating that a fraction of the bacteria-like eukaryotic genes are of alphaproteobacterial origin. Then, we show that the majority of bacteria-related eukaryotic genes actually do not point to a relationship with a specific bacterial taxonomic group. We also provide evidence that eukaryotes branch close to the last archaeal common ancestor. Our results demonstrate that there is no phylogenetic support for hypotheses involving a fusion with a bacterium other than the ancestor of mitochondria. Overall, they leave only two possible interpretations, respectively, based on the early-mitochondria hypotheses, which suppose an early endosymbiosis of an alphaproteobacterium in an archaeal host and on the slow-drip autogenous hypothesis, in which early eukaryotic ancestors were particularly prone to horizontal gene transfers.
Keywords: archaea; eukaryogenesis; evolution; horizontal gene transfer; phylogeny; tree of life.
Figures




References
-
- Albers S-V, Meyer BH. The archaeal cell envelope. Nat Rev Microbiol. 2011;9:414–426. - PubMed
-
- Allers T, Mevarech M. Archaeal genetics—the third way. Nat Rev Genet. 2005;6:58–73. - PubMed
-
- Atteia A, Adrait A, Brugiere S, Tardif M, van Lis R, Deusch O, Dagan T, Kuhn L, Gontero B, Martin W, et al. A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the α-proteobacterial mitochondrial ancestor. Mol Biol Evol. 2009;26:1533–1548. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

