RNA Interference by Single- and Double-stranded siRNA With a DNA Extension Containing a 3' Nuclease-resistant Mini-hairpin Structure
- PMID: 24399205
- PMCID: PMC3894584
- DOI: 10.1038/mtna.2013.68
RNA Interference by Single- and Double-stranded siRNA With a DNA Extension Containing a 3' Nuclease-resistant Mini-hairpin Structure
Abstract
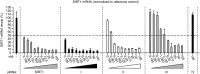
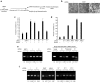
Selective gene silencing by RNA interference (RNAi) involves double-stranded small interfering RNA (ds siRNA) composed of single-stranded (ss) guide and passenger RNAs. siRNA is recognized and processed by Ago2 and C3PO, endonucleases of the RNA-induced silencing complex (RISC). RISC cleaves passenger RNA, exposing the guide RNA for base-pairing with its homologous mRNA target. Remarkably, the 3' end of passenger RNA can accommodate a DNA extension of 19-nucleotides without loss of RNAi function. This construct is termed passenger-3'-DNA/ds siRNA and includes a 3'-nuclease-resistant mini-hairpin structure. To test this novel modification further, we have now compared the following constructs: (I) guide-3'-DNA/ds siRNA, (II) passenger-3'-DNA/ds siRNA, (III) guide-3'-DNA/ss siRNA, and (IV) passenger-3'-DNA/ss siRNA. The RNAi target was SIRT1, a cancer-specific survival factor. Constructs I-III each induced selective knock-down of SIRT1 mRNA and protein in both noncancer and cancer cells, accompanied by apoptotic cell death in the cancer cells. Construct IV, which lacks the SIRT1 guide strand, had no effect. Importantly, the 3'-DNA mini-hairpin conferred nuclease resistance to constructs I and II. Resistance required the double-stranded RNA structure since single-stranded guide-3'-DNA/ss siRNA (construct III) was susceptible to serum nucleases with associated loss of RNAi activity. The potential applications of 3'-DNA/siRNA constructs are discussed.Molecular Therapy-Nucleic Acids (2014) 2, e141; doi:10.1038/mtna.2013.68; published online 7 January 2014.
Figures



References
-
- Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. - PubMed
-
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. - PubMed
-
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. - PubMed
-
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340–349. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

