Sensing microbial RNA in the cytosol
- PMID: 24400006
- PMCID: PMC3872322
- DOI: 10.3389/fimmu.2013.00468
Sensing microbial RNA in the cytosol
Abstract
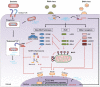
The innate immune system faces the difficult task of keeping a fine balance between sensitive detection of microbial presence and avoidance of autoimmunity. To this aim, key mechanisms of innate responses rely on isolation of pathogens in specialized subcellular compartments, or sensing of specific microbial patterns absent from the host. Efficient detection of foreign RNA in the cytosol requires an additional layer of complexity from the immune system. In this particular case, innate sensors should be able to distinguish self and non-self molecules that share several similar properties. In this review, we discuss this interplay between cytosolic pattern recognition receptors and the microbial RNA they detect. We describe how microbial RNAs gain access to the cytosol, which receptors they activate and counter-strategies developed by microorganisms to avoid this response.
Keywords: DExD/H-box helicases; RIG-I-like receptors; RNA helicases; cytosol; innate immune escape; pathogen-associated molecular patterns; pattern recognition receptors.
Figures
Similar articles
-
Cytosolic Sensors for Pathogenic Viral and Bacterial Nucleic Acids in Fish.Int J Mol Sci. 2020 Oct 2;21(19):7289. doi: 10.3390/ijms21197289. Int J Mol Sci. 2020. PMID: 33023222 Free PMC article. Review.
-
Master sensors of pathogenic RNA - RIG-I like receptors.Immunobiology. 2013 Nov;218(11):1322-35. doi: 10.1016/j.imbio.2013.06.007. Epub 2013 Jul 1. Immunobiology. 2013. PMID: 23896194 Free PMC article. Review.
-
DExD/H-box RNA helicases as mediators of anti-viral innate immunity and essential host factors for viral replication.Biochim Biophys Acta. 2013 Aug;1829(8):854-65. doi: 10.1016/j.bbagrm.2013.03.012. Epub 2013 Apr 6. Biochim Biophys Acta. 2013. PMID: 23567047 Free PMC article. Review.
-
Detection of Microbial Infections Through Innate Immune Sensing of Nucleic Acids.Annu Rev Microbiol. 2018 Sep 8;72:447-478. doi: 10.1146/annurev-micro-102215-095605. Annu Rev Microbiol. 2018. PMID: 30200854 Review.
-
RNAs Containing Modified Nucleotides Fail To Trigger RIG-I Conformational Changes for Innate Immune Signaling.mBio. 2016 Sep 20;7(5):e00833-16. doi: 10.1128/mBio.00833-16. mBio. 2016. PMID: 27651356 Free PMC article.
Cited by
-
Understanding nucleic acid sensing and its therapeutic applications.Exp Mol Med. 2023 Nov;55(11):2320-2331. doi: 10.1038/s12276-023-01118-6. Epub 2023 Nov 9. Exp Mol Med. 2023. PMID: 37945923 Free PMC article. Review.
-
Type I interferons in bacterial infections: taming of myeloid cells and possible implications for autoimmunity.Front Immunol. 2014 Sep 11;5:431. doi: 10.3389/fimmu.2014.00431. eCollection 2014. Front Immunol. 2014. PMID: 25309533 Free PMC article. Review.
-
Adeno-associated viral vector-mediated immune responses: Understanding barriers to gene delivery.Pharmacol Ther. 2020 Mar;207:107453. doi: 10.1016/j.pharmthera.2019.107453. Epub 2019 Dec 11. Pharmacol Ther. 2020. PMID: 31836454 Free PMC article. Review.
-
Effects of ionizing radiation on retinoic acid-inducible gene-I-like receptors.Biomed Rep. 2015 Jan;3(1):59-62. doi: 10.3892/br.2014.377. Epub 2014 Nov 7. Biomed Rep. 2015. PMID: 25469248 Free PMC article.
-
Single- and double-stranded viral RNA generate distinct cytokine and antiviral responses in human fetal membranes.Mol Hum Reprod. 2014 Jul;20(7):701-8. doi: 10.1093/molehr/gau028. Epub 2014 Apr 9. Mol Hum Reprod. 2014. PMID: 24723465 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

