A neutralization epitope in the hepatitis C virus E2 glycoprotein interacts with host entry factor CD81
- PMID: 24400084
- PMCID: PMC3882236
- DOI: 10.1371/journal.pone.0084346
A neutralization epitope in the hepatitis C virus E2 glycoprotein interacts with host entry factor CD81
Abstract
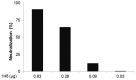
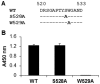
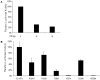
The identification of a specific immunogenic candidate that will effectively activate the appropriate pathway for neutralizing antibody production is fundamental for vaccine design. By using a monoclonal antibody (1H8) that neutralizes HCV in vitro, we have demonstrated here that 1H8 recognized an epitope mapped between residues A524 and W529 of the E2 protein. We also found that the epitope residues A524, P525, Y527 and W529 were crucial for antibody binding, while the residues T526, Y527 and W529 within the same epitope engaged in the interaction with the host entry factor CD81. Furthermore, we detected "1H8-like" antibodies, defined as those with amino acid-specificity similar to 1H8, in the plasma of patients with chronic HCV infection. The time course study of plasma samples from Patient H, a well-characterized case of post-transfusion hepatitis C, showed that "1H8-like" antibodies could be detected in a sample collected almost two years after the initial infection, thus confirming the immunogenicity of this epitope in vivo. The characterization of this neutralization epitope with a function in host entry factor CD81 interaction should enhance our understanding of antibody-mediated neutralization of HCV infections.
Conflict of interest statement
Figures




References
-
- Centers for Disease Control and Prevention (2012) CDC Health Information for International Travel (Oxford Univ Press, New York).
-
- Lauer GM, Walker BD (2001) Hepatitis C virus infection. N Engl J Med 345: 41–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

